Recent advances in single-cell sequencing technologies have provided unprecedented opportunities to measure the gene expression profile and RNA velocity of individual cells. However, modeling transcriptional dynamics is computationally challenging because of the high-dimensional, sparse nature of the single-cell gene expression measurements and the nonlinear regulatory relationships.
Researchers at Yale University and the University of California, Irvine have developed DeepVelo, a neural network-based ordinary differential equation that can model complex transcriptome dynamics by describing continuous-time gene expression changes within individual cells. The researchers applied DeepVelo to public datasets from different sequencing platforms to (i) formulate transcriptome dynamics on different time scales, (ii) measure the instability of cell states, and (iii) identify developmental driver genes via perturbation analysis. Benchmarking against the state-of-the-art methods showed that DeepVelo can learn a more accurate representation of the velocity field. Furthermore, perturbation studies revealed that single-cell dynamical systems could exhibit chaotic properties. In summary, DeepVelo allows data-driven discoveries of differential equations that delineate single-cell transcriptome dynamics.
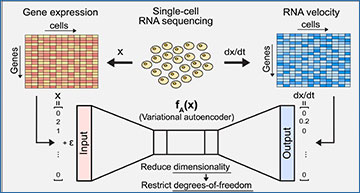
Schematic for DeepVelo
(A) Gene expression profiles and corresponding transcriptional velocities can be derived from scRNA-seq. After learning the mapping between gene expression and RNA velocity, the VAE represents a neural differential equation that encapsulates transcriptome dynamics. (B) Given an initial condition and time, our framework solves for the future gene expression state by integrating the VAE with a black-box ODE solver. (C) Our approach can simulate trajectories to evaluate the instability of cell states in a dynamical system. (D) DeepVelo can perform in silico perturbation studies to identify the developmental driver genes that determine the fate of cell bifurcations. (E) Comparison between DeepVelo and existing single-cell methods.
Chen Z, King WC, Hwang A, Gerstein M, Zhang J. (2022) DeepVelo: Single-cell transcriptomic deep velocity field learning with neural ordinary differential equations. Sci Adv 8(48):eabq3745. [article]