Machine learning methods, particularly neural networks trained on large datasets, are transforming how scientists approach scientific discovery and experimental design. However, current state-of-the-art neural networks are limited by their uninterpretability: Despite their excellent accuracy, they cannot describe how they arrived at their predictions. Here, using an “interpretable-by-design” approach, New York University researchers have developed a neural network model that provides insights into RNA splicing, a fundamental process in the transfer of genomic information into functional biochemical products. Although they designed the model to emphasize interpretability, its predictive accuracy is on par with state-of-the-art models. To demonstrate the model’s interpretability, the researchers introduce a visualization that, for any given exon, allows them to trace and quantify the entire decision process from input sequence to output splicing prediction. Importantly, the model revealed uncharacterized components of the splicing logic, which they experimentally validated. This study highlights how interpretable machine learning can advance scientific discovery.
Data generation and interpretable-by-design machine learning model
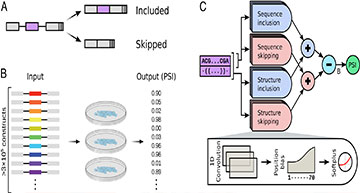
(A) All reporters in the assay share the same three-exon design and differ only in their middle exon, which contains a random 70-nucleotide-long sequence. Depending on its sequence, an exon might be included, skipped, or a probabilistic mix of the two. Each reporter includes a unique barcode at the end of the third exon so that exon identity can be inferred in exon-skipping products. (B) The assay includes over 3×105 different reporters. The reporters were transfected into HeLa cells in a pooled fashion in three biological replicates. High-throughput sequencing then provides a “percent spliced in” (PSI) value to each reporter. (C) The machine learning model consists of both short convolution filters (applied to exon sequence only) and long convolution filters (applied to both exon sequence and predicted structure). The output of these filters (strength) can depend on the position along the exon. Half of the filters are designated as inclusion filters, and the rest are skipping filters. Predicted PSI is computed from the difference between the total strength of inclusion filters and the total strength of skipping filters, after adding an initial basal strength (B).
Availability – https://github.com/regev-lab/interpretable-splicing-model
Liao SE, Sudarshan M, Regev O. (2023) Deciphering RNA splicing logic with interpretable machine learning. PNAS 120(41):e2221165120. [article]