Differential allele-specific expression (ASE) is a powerful tool to study context-specific cis-regulation of gene expression. Such effects can reflect the interaction between genetic or epigenetic factors and a measured context or condition. Single-cell RNA sequencing (scRNA-seq) allows the measurement of ASE at individual-cell resolution, but there is a lack of statistical methods to analyze such data. Johns Hopkins University researchers have developed Differential Allelic Expression using Single-Cell data (DAESC), a powerful method for differential ASE analysis using scRNA-seq from multiple individuals, with statistical behavior confirmed through simulation. DAESC accounts for non-independence between cells from the same individual and incorporates implicit haplotype phasing. Application to data from 105 induced pluripotent stem cell (iPSC) lines identifies 657 genes dynamically regulated during endoderm differentiation, with enrichment for changes in chromatin state. Application to a type-2 diabetes dataset identifies several differentially regulated genes between patients and controls in pancreatic endocrine cells. DAESC is a powerful method for single-cell ASE analysis and can uncover novel insights on gene regulation.
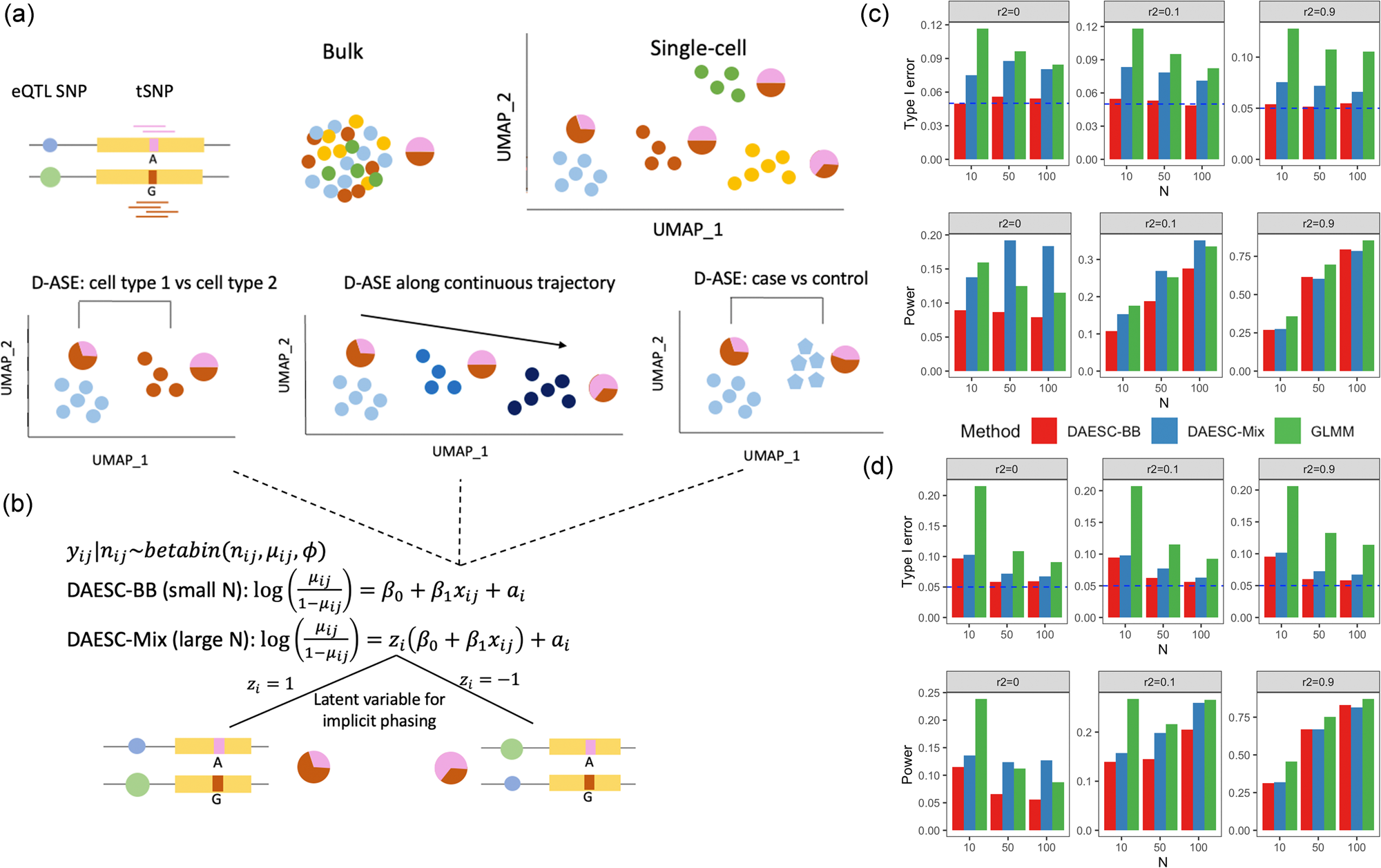
Schematic of DAESC and simulation studies
a Schematic of allele-specific expression (ASE) measured in bulk tissue and single cells, and three types of differential ASE analysis. Pie charts represent the relative expression of two alleles of the transcribed SNP (tSNP). b DAESC models. DAESC accounts for sample repeat structure (multiple cells per sample) using random effects {a}_{i} and implicit haplotype phasing using latent variables {z}_{i}. c, d Type I error and power observed in simulation studies for differential ASE, c along a continuous cell state and d binary case–control disease status. Type I error and power are computed under nominal significance threshold P < 0.05 (no multiple comparison adjustment). Likelihood ratio test is used for DAESC-BB and DAESC-Mix and z-test is used for GLMM. All tests are two-sided. Allele-specific read counts are simulated from beta-binomial mixture model assuming only one eQTL drives ASE at a tSNP. The linkage disequilibrium between the eQTL and the tSNP is varied to {r}^{2}=0,0.1,0.9, and the sample size (number of individuals) is varied to N = 10, 50, 100.
Availability – The DAESC R package and other analysis scripts are available on GitHub: https://github.com/gqi/DAESC
Qi G, Strober BJ, Popp JM, Keener R, Ji H, Battle A. (2023) Single-cell allele-specific expression analysis reveals dynamic and cell-type-specific regulatory effects. Nat Commun 14(1):6317. [article]