Liquid biopsies provide a means for the profiling of cell-free RNAs secreted by cells throughout the body. Although well-annotated coding and non-coding transcripts in blood are readily detectable and can serve as biomarkers of disease, the overall diagnostic utility of the cell-free transcriptome remains unclear. Researchers from the University of California Santa Cruz show that RNAs derived from transposable elements and other repeat elements are enriched in the cell-free transcriptome of patients with cancer, and that they serve as signatures for the accurate classification of the disease.
The researchers used repeat-element-aware liquid-biopsy technology and single-molecule nanopore sequencing to profile the cell-free transcriptome in plasma from patients with cancer and to examine millions of genomic features comprising all annotated genes and repeat elements throughout the genome. By aggregating individual repeat elements to the subfamily level, they found that samples with pancreatic cancer are enriched with specific Alu subfamilies, whereas other cancers have their own characteristic cell-free RNA profile. These findings show that repetitive RNA sequences are abundant in blood and can be used as disease-specific diagnostic biomarkers.
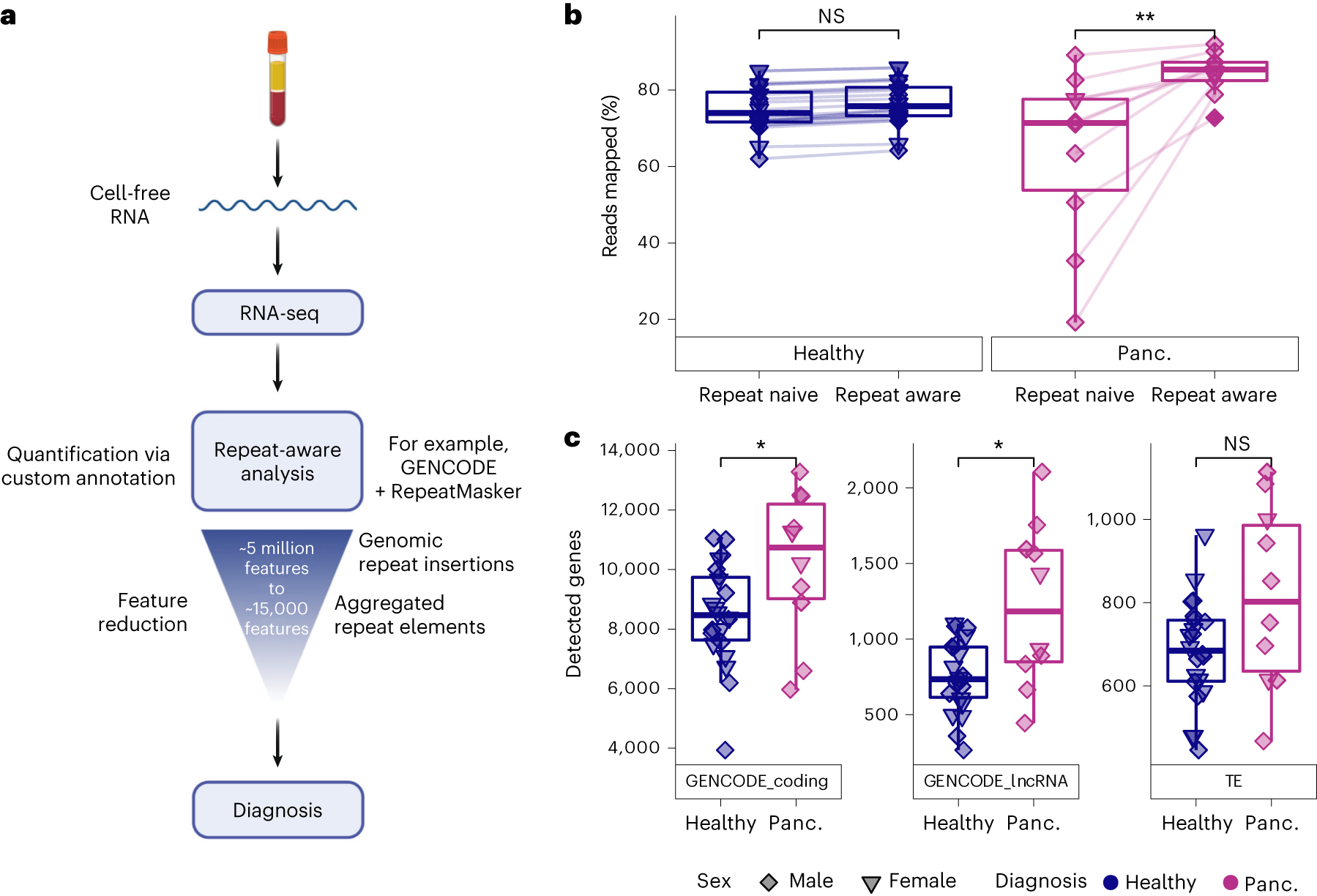
Cell-free RNA transcriptome profiling using repeat-aware COMPLETE-seq
a, Diagram of COMPLETE-seq RNA liquid-biopsy technology, highlighting the use of repeat-derived cell-free RNAs aggregated into a tractable feature set to enable diagnostic modelling. b, Comparison of mapping rates between use of a repeat-naive (GENCODE v.39) reference annotation (**P = 0.0039) and repeat-aware reference annotation (Wilcoxon, paired, two-sided). c, Comparison of gene detection distributions for each cohort across coding genes (GENCODE_coding; *P = 0.043), lncRNAs (GENCODE_lncRNA; *P = 0.035) and TE subfamilies (Wilcoxon, two-sided). For the box plots, the centre line represents the median, the box limits are upper and lower quartiles and whiskers represent 1.5× interquartile range. NS, not significant; panc., pancreatic cancer.
Availability – Custom code used in this study is available on GitHub at https://github.com/rreggiar/exRNA_disease_biomarkers.
Reggiardo RE, Maroli SV, Peddu V, Davidson AE, Hill A, LaMontagne E, Aaraj YA, Jain M, Chan SY, Kim DH. (2023) Profiling of repetitive RNA sequences in the blood plasma of patients with cancer. Nat Biomed Eng [Epub ahead of print]. [article]