In recent years, high-throughput single-cell transcriptomics and chromatin accessibility methods have generated numerous resources and provided new insights into the cell heterogeneity of tissues or whole organisms. Protocols utilizing pool-split and barcoded oligo ligation strategies greatly improve the throughput of scRNA-seq. In this work, researchers at the Zhejiang University School of Medicine have developed single-cell combinatorial hybridization sequencing (CH-seq), another high-throughput and low-cost pool-split-based method, to profile gene expression in single cells. The researchers applied CH-RNA-seq to profile over 1 million cells from 19 tissues in neotenic adult axolotls, metamorphosed adult axolotls and larval stage axolotls. Previous efforts of axolotl tissue-based bulk RNA-seq (TRIzol extraction) and scRNA-seq identified tissue specific transcripts related to limb regeneration and human diseases. Based on these efforts, the adult axolotl cell landscape, created using CH-seq to detect transcripts fixed in dissociated cells, comprehensively covered cell diversity within and between axolotl tissues at single-cell level. The single-cell transcriptomic comparison between neotenic and metamorphosed axolotls allowed the researchers to explore perturbed genes and cell type in metamorphosis.
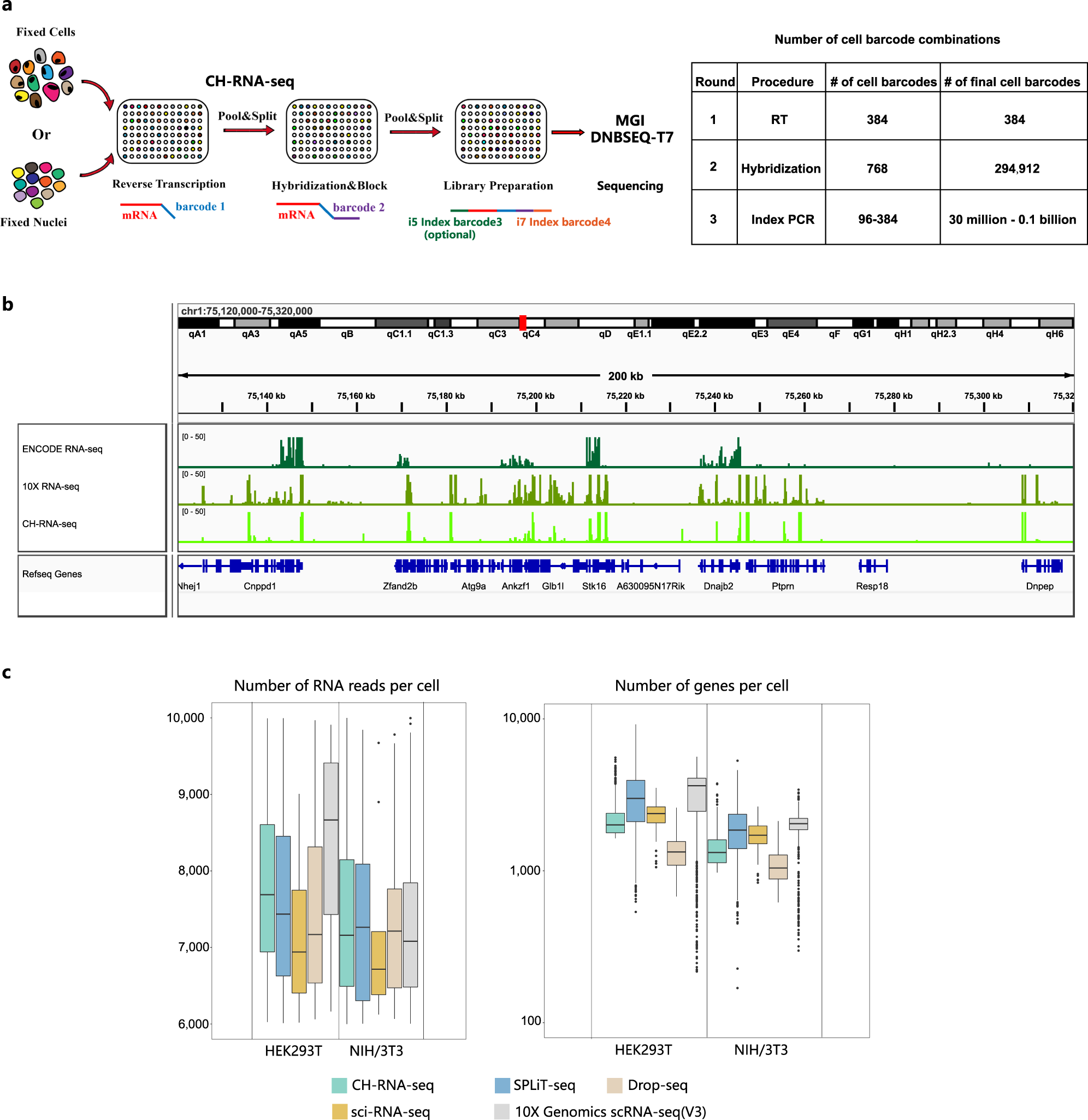
Profiling of the transcriptome in single cells using CH-seq
a Diagram illustrating the experimental workflow for combinatorial hybridization sequencing b Representative genome browser view of CH-seq NIH/3T3 cell data, 10X Genomics data and ENCODE NIH/3T3 cell data read coverage using an integrative genomics viewer (IGV). ENCODE datasets were obtained from the Gene Expression Omnibus with the accession numbers mentioned above. c Box plots showing the number of uniquely mapped RNA reads and the number of genes detected per cell from HEK293T and NIH/3T3 cells (n = 842 cells, The boxplots are defined by the 25th and 75th percentiles, with the centre as the median, the minima and maxima extend to the largest value until 1.5 of the interquartile range and the smallest value at most 1.5 of interquartile range, respectively.). Original version of the number of genes and reads in sci-RNA-seq, SPLiT-seq, Drop-seq and “10X Genomics scRNA-seq” were obtained as source data in Paired-seq. The original dataset could be obtained from the Gene Expression Omnibus with the accession numbers mentioned above.
Ye F, Zhang G, E W, Chen H, Yu C, Yang L, Fu Y, Li J, Fu S, Sun Z, Fei L, Guo Q, Wang J, Xiao Y, Wang X, Zhang P, Ma L, Ge D, Xu S, Caballero-Pérez J, Cruz-Ramírez A, Zhou Y, Chen M, Fei JF, Han X, Guo G. (2022) Construction of the axolotl cell landscape using combinatorial hybridization sequencing at single-cell resolution. Nat Commun 13(1):4228. [article]