The current lack of benchmark datasets with inbuilt ground-truth makes it challenging to compare the performance of existing long-read isoform detection and differential expression analysis workflows. Researchers from the Walter and Eliza Hall Institute of Medical Research present a benchmark experiment using two human lung adenocarcinoma cell lines that were each profiled in triplicate together with synthetic, spliced, spike-in RNAs (“sequins”). Samples were deeply sequenced on both Illumina short-read and Oxford Nanopore Technologies long-read platforms. Alongside the ground-truth available via the sequins, the researchers created in silico mixture samples to allow performance assessment in the absence of true positives or true negatives. These results show that, StringTie2 and bambu outperformed other tools from the 6 isoform detection tools tested, DESeq2, edgeR and limma-voom were best amongst the 5 differential transcript expression tools tested and there was no clear front-runner for performing differential transcript usage analysis between the 5 tools compared, which suggests further methods development is needed for this application.
Overview of the experimental design and benchmark analysis
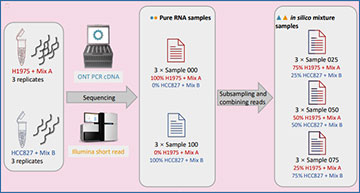
(a) Summary of the experimental design involving pure RNA samples obtained from 2 cancer cell line (H1975 and HCC827) with 2 synthetic sequins spike-in control mixes (A and B) and in silico generated mixture samples. (b) Overview of the analysis workflow to benchmark the performance of different RNA-seq analysis tools for isoform detection and quantification, differential transcript expression (DTE) and differential transcript usage (DTU). Analysis steps and selected methods are shown in shaded boxes with solid borders, while evaluation metrics are listed in boxes with dashed borders.
Dong X, Du MRM, Gouil Q, Tian L, Baldoni PL, Smyth GK, Amarasinghe SL, Law CW, Ritchie ME. (2022) Benchmarking long-read RNA-sequencing analysis tools using in silico mixtures. bioRXiv [online preprint]. [abstract]