In single-cell RNA sequencing (scRNA-seq) data analysis, addressing batch effects – technical artifacts stemming from factors such as varying sequencing technologies, equipment, and capture times – is crucial. These factors cause unwanted variation in the data and often obfuscate the underlying biological signal of interest. The Joint and Individual Variation Explained (JIVE) method can be used to extract shared biological patterns from multi-source sequencing data while adjusting for individual non-biological variations (i.e., batch effect). However, its current implementation is originally designed for bulk sequencing data, making it computationally infeasible for large-scale single-cell sequencing datasets.
Researchers at Miami University enhanced JIVE for large-scale scRNA-seq data by boosting its computational efficiency and tailoring it to the single-cell context. Additionally, the researchers introduce a novel application of JIVE which they use to perform batch-effect correction on multiple scRNA-seq datasets. Thier enhanced JIVE method aims to decompose scRNA-seq datasets into a joint structure capturing the true biological variability and individual structures which capture technical variability within each batch. This joint structure is then suitable for use in downstream analyses. The researchers employed four evaluation metrics and benchmarked the results against two other popular tools, Seurat v3 and Harmony, which were developed for this purpose. They found that JIVE performed best in metrics that consider local neighborhoods (kBET and LISI) and in scenarios in which the original data contained distinct differences between batches and cell types.
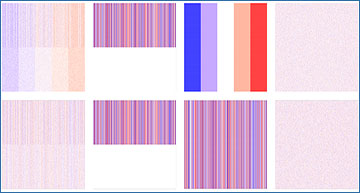
Heatmaps for simulation data used in JIVE benchmarks
The first row shows the final data matrix A and the second row shows the final data matrix B and their respective decompositions. The first column is the data used as input into the JIVE algorithm. These final datasets were created by adding the second column representing the same joint structure shared between datasets, the third column representing the individual structure unique to each dataset, and the fourth column representing white noise.
Availability – The JIVE implementation used for this analysis can be found at https://github.com/oconnell-statistics-lab/scJIVE.
Hastings J, Lee D, O’Connell MJ. (2023) Batch-effect correction in single-cell RNA sequencing data using JIVE. bioRXiv [online preprint]. [abstract]