There has been significant progress made in the field of nanopore biosensor development and sequencing applications, which address previous limitations that restricted widespread nanopore use. These innovations, paired with the large-scale commercialization of biological nanopore sequencing by Oxford Nanopore Technologies, are making the platforms a mainstay in contemporary research laboratories. Equipped with the ability to provide long- and short read sequencing information, with quick turn-around times and simple sample preparation, nanopore sequencers are rapidly improving our understanding of unsolved genetic, transcriptomic, and epigenetic problems. However, there remain some key obstacles that have yet to be improved. Researchers from the University of New Mexico provide a general introduction to nanopore sequencing principles, discussing biological and solid-state nanopore developments, obstacles to single-base detection, and library preparation considerations. They present examples of important clinical applications to give perspective on the potential future of nanopore sequencing in the field of molecular diagnostics.
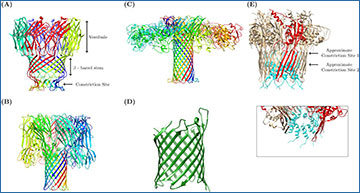
Anatomy of a Nanopore. A depiction of biological nanopores employed in sequencing
(A) MspA pore (PDB ID 1UUN) with a constriction site diameter of 1.2 nm, and a stem length of ~3.7 nm (B) Alpha Hemolysin (PDB ID 7AHL) with a constriction site diameter of 2.6 nm and a stem length of 5.2 nm (C) Aerolysin porin (PDB ID 5JZT) with a constriction site diameter of 1 nm, and a stem length ~10 nm. (D) A simplified depiction of OmpG pore (PDB ID 2F1C) with a constriction site of 1.3 nm. (E) CsgG-CsgF mutant (PDB ID 6SI7) with two constriction sites: the original CsgG constriction of 1 nm diameter (chain monomer of the original CsgG pore is depicted in red); and a secondary constriction caused by the insertion of CsgF (Cyan residues) with a 1.5 nm diameter. The insert shows the approximated diameter of the second constriction site, ~1.5 nm. Protein chains are depicted in different colors to help with distinction. All proteins were recreated in Chimera utilizing PDB IDs from published protein structures (noted PDB ID numbers).
MacKenzie M, Argyropoulos C. (2023) An Introduction to Nanopore Sequencing: Past, Present, and Future Considerations. Micromachines 14(2), 459. [article]