A team led by researchers at the Ohio State University have developed a novel positive false discovery rate (pFDR) controlling method for testing gene-specific hypotheses using a gene-specific covariate variable, such as gene length. The researchers suppose the null probability depends on the covariate variable. In this context, they propose a rejection rule that accounts for heterogeneity among tests by employing two distinct types of null probabilities. They establish a pFDR estimator for a given rejection rule by following Storey’s q-value framework. A condition on a type 1 error posterior probability is provided that equivalently characterizes our rejection rule. The researchers also present a suitable procedure for selecting a tuning parameter through cross-validation that maximizes the expected number of hypotheses declared significant. A simulation study demonstrates that this method is comparable to or better than existing methods across realistic scenarios. In data analysis, they find support for their method’s premise that the null probability varies with a gene-specific covariate variable.

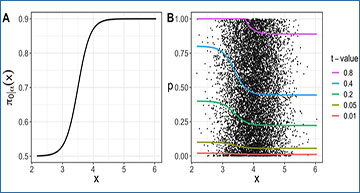
An example function π0|α(x) is depicted in Figure A, and the rejection regions’ upper bounds created by five distinct t-values are illustrated in Figure B.
Jeon H, Lim KS, Nguyen Y, Nettleton D. (2023) Adjusting for gene-specific covariates to improve RNA-seq analysis. Bioinformatics [Epub ahead of print]. [abstract]