The ever-growing compendium of genetic variants associated with human pathologies demands new methods to study genotype-phenotype relationships in complex tissues in a high-throughput manner. ETH Zurich researchers have developed adeno-associated virus (AAV)-mediated direct in vivo single-cell CRISPR screening, termed AAV-Perturb-seq, a tuneable and broadly applicable method for transcriptional linkage analysis as well as high-throughput and high-resolution phenotyping of genetic perturbations in vivo. The researchers applied AAV-Perturb-seq using gene editing and transcriptional inhibition to systematically dissect the phenotypic landscape underlying 22q11.2 deletion syndrome genes in the adult mouse brain prefrontal cortex. They identified three 22q11.2-linked genes involved in known and previously undescribed pathways orchestrating neuronal functions in vivo that explain approximately 40% of the transcriptional changes observed in a 22q11.2-deletion mouse model. These findings suggest that the 22q11.2-deletion syndrome transcriptional phenotype found in mature neurons may in part be due to the broad dysregulation of a class of genes associated with disease susceptibility that are important for dysfunctional RNA processing and synaptic function.
In vivo single-nucleus pooled CRISPR screening in the adult brain enabled by systemic administration of AAV.PHP.B and 5′ gRNA capture
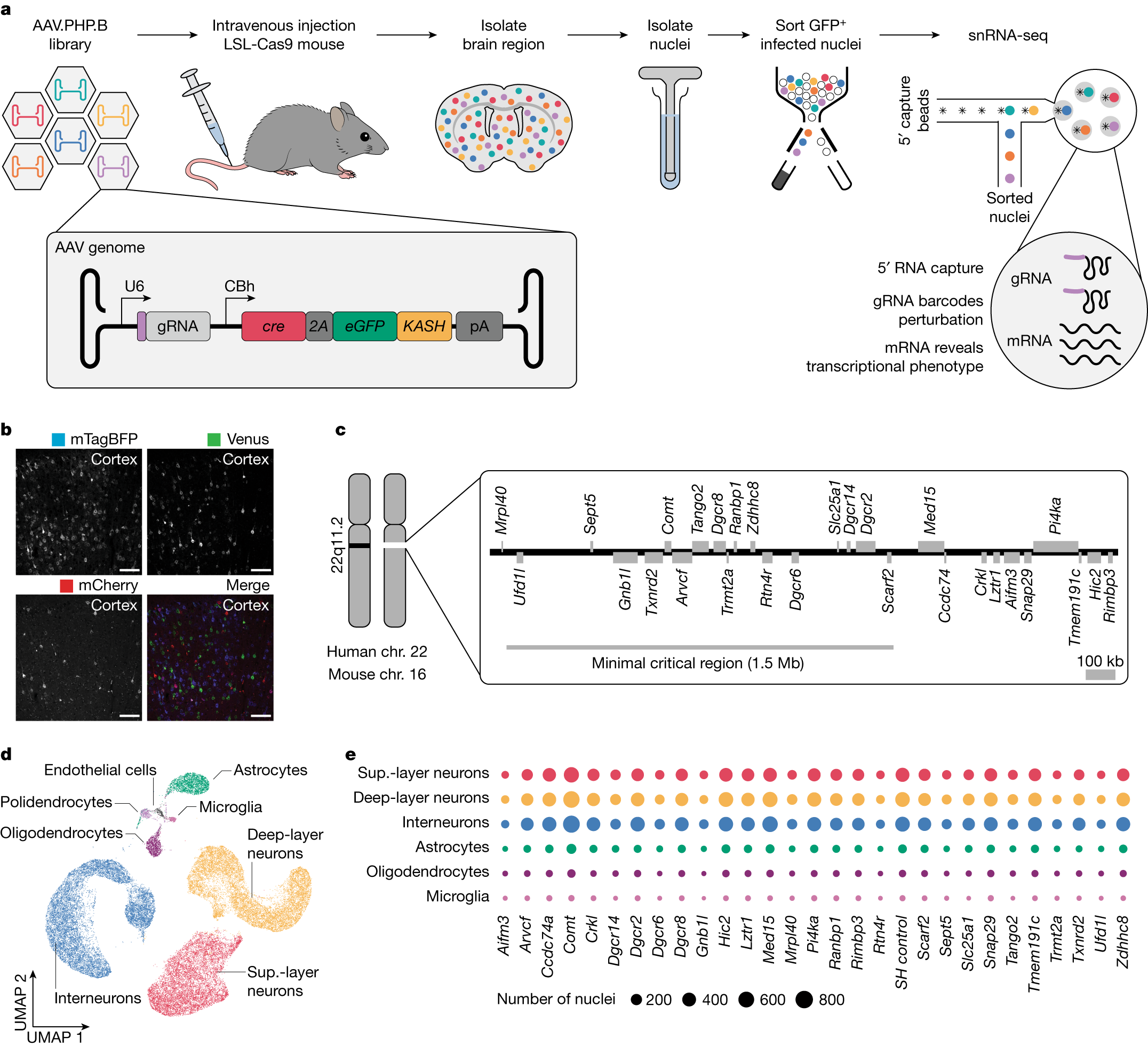
a, The AAV-Perturb-seq experimental pipeline. b, Expression of mTagBFP, Venus and mCherry in the prefrontal cortex after systemic injection of an equal mixture of 5.0 × 109 total AAV particles. Scale bars, 100 µm. The experiments were repeated in n = 3 mice. c, Representation of the 22q11.2 locus showing the genes expressed in the adult mouse prefrontal cortex. The human 22q11.2 locus is conserved in mouse chromosome (chr.) 16. d, UMAP embedding of around 150,000 AAV.PHP.B-infected nuclei isolated from the mouse prefrontal cortex. e, The number of nuclei with a unique gRNA for each perturbation across cell types.
This study establishes a flexible and scalable direct in vivo method to facilitate causal understanding of biological and disease mechanisms with potential applications to identify genetic interventions and therapeutic targets for treating disease.
Availability – Custom made scripts are available at the Platt laboratory GitHub (https://github.com/plattlab/AAV-Perturb-seq).
Santinha AJ, Klingler E, Kuhn M, Farouni R, Lagler S, Kalamakis G, Lischetti U, Jabaudon D, Platt RJ. (2023) Transcriptional linkage analysis with in vivo AAV-Perturb-seq. Nature [Epub ahead of print]. [article]