University of Virginia School of Medicine researchers and their collaborators have created a powerful new tool they say will benefit essentially every area of biomedical research, from understanding how healthy cells function to the battle against cancer and neurodegenerative diseases such as Alzheimer’s.
UVA’s Gloria M. Sheynkman, PhD, and her team have developed a new and better way to identify proteins our genes make. These proteins are workhorses inside our cells, playing critical roles in both normal cellular functions and in the development of diseases. Better understanding these proteins will help scientists unlock the mysteries of how our cells work and how diseases take hold, and targeting the proteins could lead to important new treatments for a wide variety of conditions.
“Genes don’t drive disease states, protein isoforms, or ‘proteoforms,’ do,” said Sheynkman, of UVA’s Department of Molecular Physiology and Biological Physics. Erin Jeffery, PhD, a senior scientist in Sheynkman’s lab, noted, “To discover disease-relevant protein isoforms, the fields of genomics [the study of genomes]and proteomics [the study of proteins]need each other’s technology to move disease research forward. In our lab we are using long-read RNA sequencing combined with mass spectrometry as a bridge to bring the two fields together.”
Powerful Proteins
Understanding cellular proteins can be tremendously challenging for scientists. Our genes often manufacture proteins that have similar functions but are ever so slightly different. These variations, known as “isoforms,” add great complexity to the already complex process of interpreting cellular processes. You might think of understanding isoforms a bit like choosing a car – your options may appear to be similar at first, with four wheels and an engine, but they can vary greatly in features and fuel efficiency. It’s important to understand those distinctions.
Scientists now commonly use a process called “mass spectrometry” to understand isoforms. But this chemical analysis has limitations, and there are a tremendous number of protein isoforms to identify – estimates suggest there may be more than 300,000.
Sheynkman’s new tool takes a two-pronged approach, combining mass spectrometry data with a technology called long-read RNA sequencing that is used to predict isoforms. To develop this new approach, the researchers had to create an algorithm to integrate the long-read sequencing results with the mass-spectrometry data. The resulting “paired data” allows researchers to identify isoforms that have, until now, defied analysis.
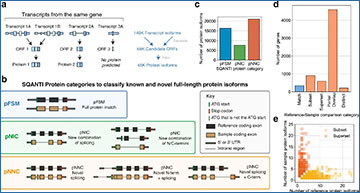
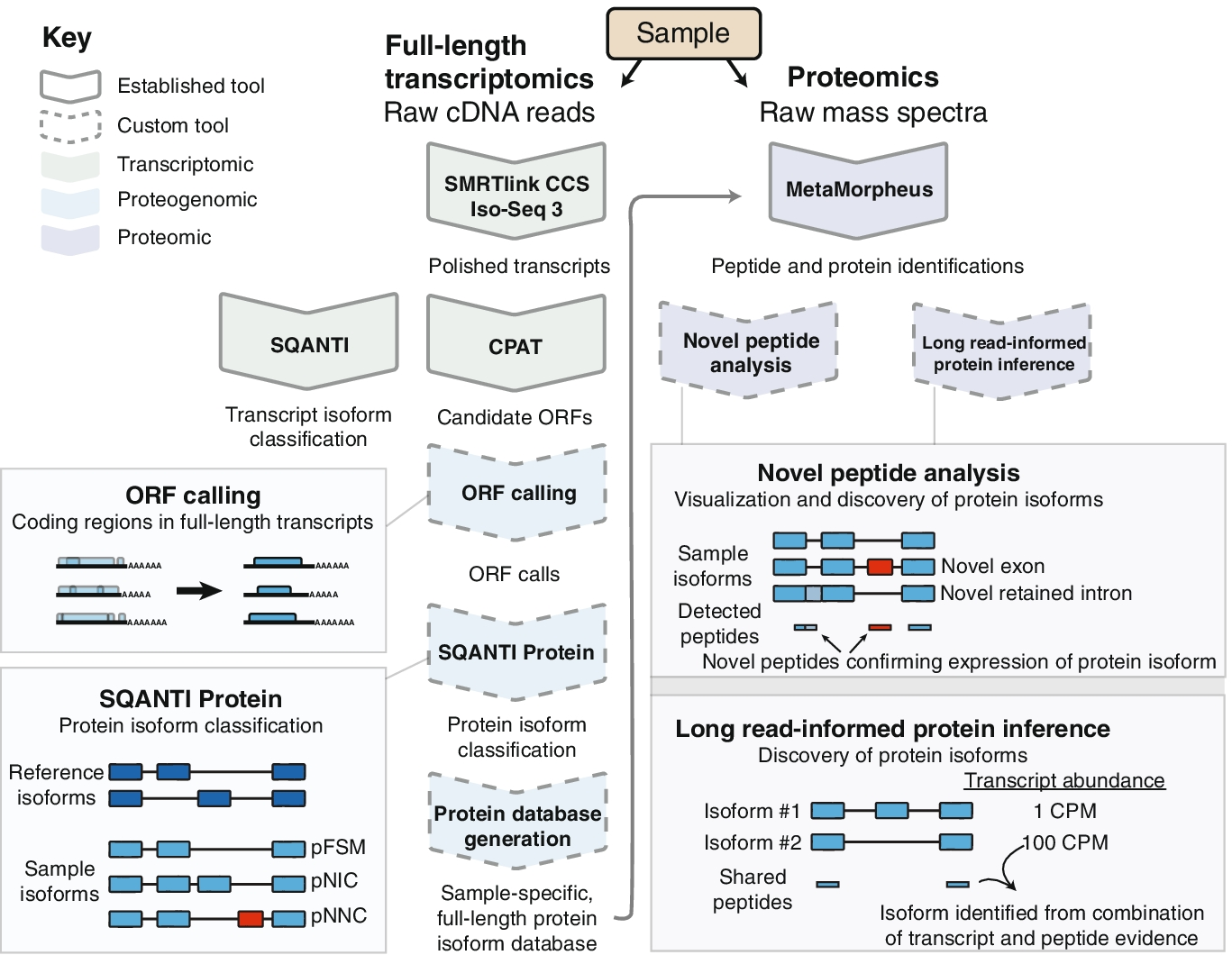
Long-read proteogenomic approach for enhanced sample-specific protein identification
Schematic of the long-read proteogenomics pipeline for improved protein isoform characterization. The pipeline includes approaches for ORF calling from long transcript reads, an automated protein isoform classification (SQANTI Protein), novel protein isoform detection, and a long-read-informed protein inference algorithm. CPM—full-length read counts per million
After successfully testing their new tool to identify thousands of unknown isoforms, the scientists have released an open-source “pipeline” that will allow other researchers to take advantage of the tool freely, for the benefit of medical research everywhere. The end result will be a much better understanding of the molecular causes of disease.
“We are very interested in application of this approach to study aberrant protein isoform expression in disease, especially complex diseases and cancer,” Sheynkman said. “Our ultimate goal is to apply proteogenomics, which at its heart is personalized proteomics, to rapidly detect patient-specific proteoforms that are driving disease pathways. We are excited about merging technologies, disciplines and new ideas to make progress in this goal.”
Source – UVA Health
Miller RM, Jordan BT, Mehlferber MM, Jeffery ED, Chatzipantsiou C, Kaur S, Millikin RJ, Dai Y, Tiberi S, Castaldi PJ, Shortreed MR, Luckey CJ, Conesa A, Smith LM, Deslattes Mays A, Sheynkman GM. (2022) Enhanced protein isoform characterization through long-read proteogenomics. Genome Biol 23(1):69. [article]