RNA-sequencing (RNA-seq) is a gold-standard method to profile genome-wide changes in gene expression. RNA-seq uses high-throughput sequencing technology to quantify the amount of RNA in a biological sample. With the increasing popularity of RNA-seq, many variations on the protocol have been proposed to extract unique and relevant information from biological samples. 3′ Tag-Seq (also called TagSeq, 3′ Tag-RNA-Seq, and Quant-Seq 3′ mRNA-Seq) is one RNA-seq variation where the 3′ end of the transcript is selected and amplified to yield one copy of cDNA from each transcript in the biological sample.
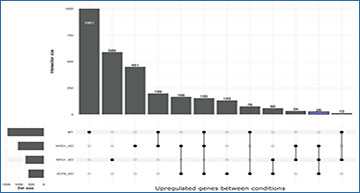
University of California, Merced researchers present a simple, easy-to-use, and publicly available computational workflow to analyze 3′ Tag-Seq data. The workflow begins by trimming sequence adapters from raw FASTQ files. The trimmed sequence reads are checked for quality using FastQC and aligned to the reference genome, and then read counts are obtained using STAR. Differential gene expression analysis is performed using DESeq2, based on differential analysis of gene count data. The outputs of this workflow are MA plots, tables of differentially expressed genes, and UpSet plots. This protocol is intended for users specifically interested in analyzing 3′ Tag-Seq data, and thus normalizations based on transcript length are not performed within the workflow. Future updates to this workflow could include custom analyses based on the gene counts table as well as data visualization enhancements.
3′ Tag-Seq analysis workflow
(i) Adapters added to raw RNA-seq reads are trimmed using BBDuk. (ii) A quality control report is generated for trimmed reads using FastQC. (iii) Reads passing the QC check are aligned to the reference genome using STAR and a gene count table is created. (iv) The gene count table is used to run differential gene expression analysis using DESeq2, and the DESeq2 output is saved as a table to a file for downstream usage.
Paropkari AD, Bapat PS, Sindi SS, Nobile CJ. (2023) A Computational Workflow for Analysis of 3′ Tag-Seq Data. Curr Protoc 3(2):e664. [article]