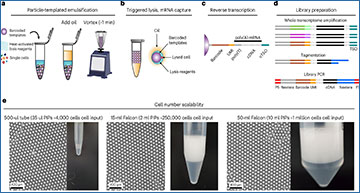
Current single-cell RNA-sequencing approaches have limitations that stem from the microfluidic devices or fluid handling steps required for sample processing. Researchers at the University of California, Berkeley have developed a method that does not require specialized microfluidic devices, expertise or hardware. This approach is based on particle-templated emulsification, which allows single-cell encapsulation and barcoding of cDNA in uniform droplet emulsions with only a vortexer. Particle-templated instant partition sequencing (PIP-seq) accommodates a wide range of emulsification formats, including microwell plates and large-volume conical tubes, enabling thousands of samples or millions of cells to be processed in minutes. The researchers demonstrate that PIP-seq produces high-purity transcriptomes in mouse-human mixing studies, is compatible with multiomics measurements and can accurately characterize cell types in human breast tissue compared to a commercial microfluidic platform. Single-cell transcriptional profiling of mixed phenotype acute leukemia using PIP-seq reveals the emergence of heterogeneity within chemotherapy-resistant cell subsets that were hidden by standard immunophenotyping. PIP-seq is a simple, flexible and scalable next-generation workflow that extends single-cell sequencing to new applications.
Rapid and scalable templated emulsification for single-cell genomics
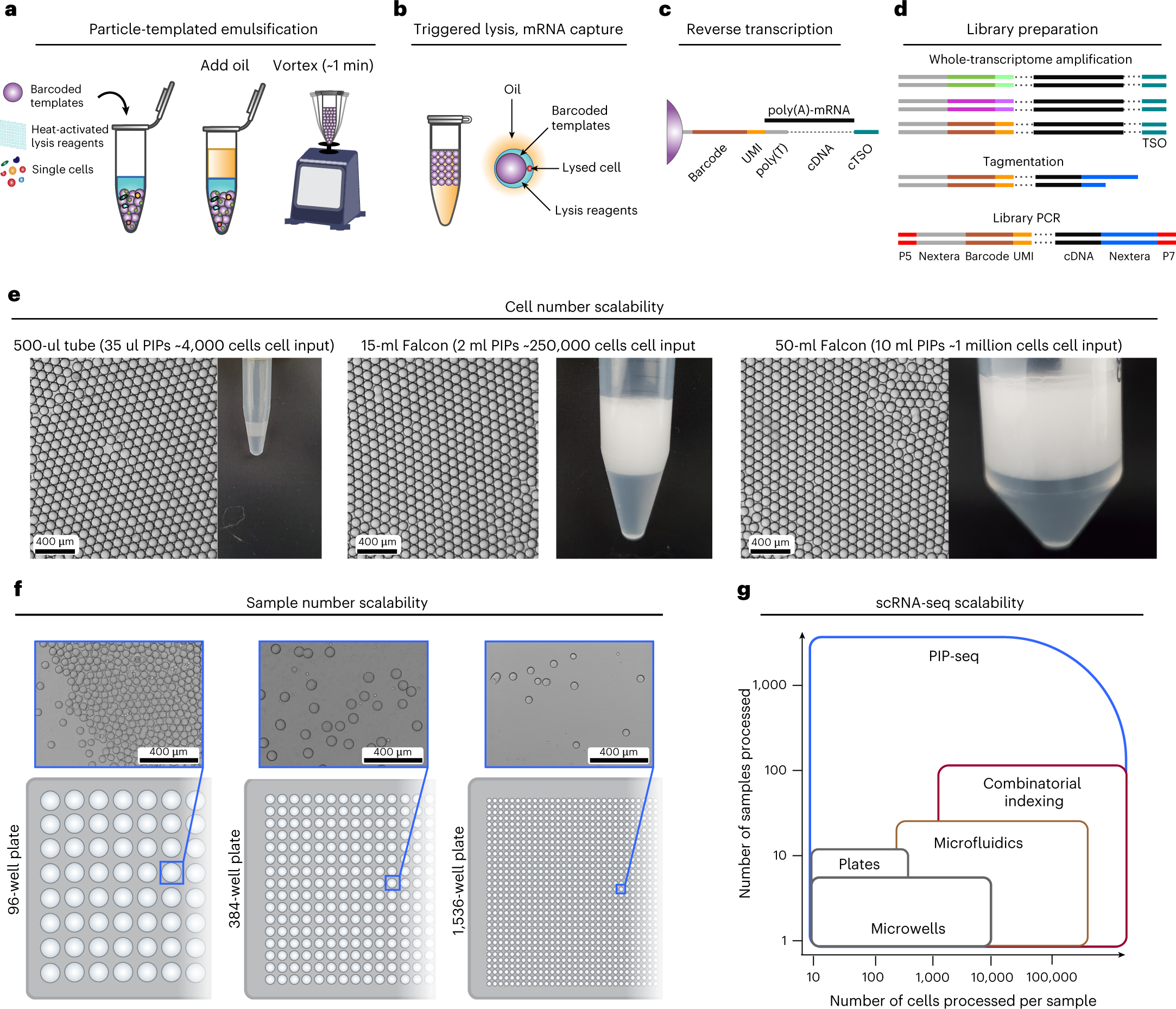
a–d, PIP-seq enables the encapsulation, lysis and barcoding of single cells. a, Schematic of the emulsification process. Barcoded particle templates, cells and lysis reagents are combined with oil and vortexed to generate monodispersed droplets. b, Heat activation of PK results in lysis and release of mRNA that is captured on bead-bound barcoded poly(T) oligonucleotides. c, Oil removal is followed by bulk reverse transcription of mRNA into cDNA. cTSO is the complement of the template switch oligonucleotide. d, Barcoded whole-transcriptome-amplified cDNA is prepared for Illumina sequencing. e–g, Efficient single-bead, single-drop encapsulation at scale. e, Particle-templated emulsification in different-sized tubes (1.5 ml, 15 ml and 50 ml) produces monodispersed emulsions capable of barcoding orders of magnitude different cell numbers. f, PIP-seq is compatible with plate-based emulsification, including 96-, 384- and 1,536-well plate formats. Representative images are shown from experiments completed three times. g, The estimated ability of different technologies to easily scale with respect to cell and sample number.
Clark IC, Fontanez KM, Meltzer RH, Xue Y, Hayford C, May-Zhang A, D’Amato C, Osman A, Zhang JQ, Hettige P, Ishibashi JSA, Delley CL, Weisgerber DW, Replogle JM, Jost M, Phong KT, Kennedy VE, Peretz CAC, Kim EA, Song S, Karlon W, Weissman JS, Smith CC, Gartner ZJ, Abate AR. (2023) Microfluidics-free single-cell genomics with templated emulsification. Nat Biotechnol [Epub ahead of print]. [article]