Over the last decade, single-cell sequencing has transformed many fields. It has enabled the unbiased molecular phenotyping of even whole organisms with unprecedented cellular resolution. In the field of human genetics, where the phenotypic consequences of genetic and epigenetic alterations are of central concern, this transformative technology promises to functionally annotate every region in the human genome and all possible variants within them at a massive scale.
In this review aimed at the clinicians in human genetics, researchers from the University Hospital Schleswig-Holstein describe the current status of the field of single-cell sequencing and its role for human genetics, including how the technology works as well as how it is being applied to characterize and monitor diseases, to develop human cell atlases, and to annotate the genome.
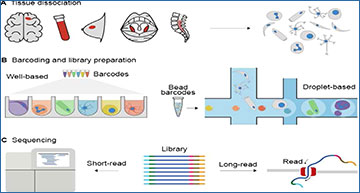
The experimental workflow of sc-seq
(A) The workflow starts with the dissociation of the biopsy samples into a cellular or nuclear suspension. (B) This is followed by cellular barcoding using droplet- or well-based technologies that enables pooled sequencing of the molecules from all the cells. (C) The final step is sequencing. Although long-read sequencing is rarely used in current analysis workflows, it has potential for identification of splice variants in the case of sc-transcriptome-seq or structural variants in the case of sc-genome-seq as well as to reduce sequencing costs
Sreenivasan VKA, Henck J, Spielmann M. (2022) Single-cell sequencing: promises and challenges for human genetics. Medizinische Genetik 34(4); 261-273. [article]