Single-cell RNA sequencing (scRNA-seq) continues to expand our knowledge by facilitating the study of transcriptional heterogeneity at the level of single cells. Despite this technology’s utility and success in biomedical research, technical artifacts are present in scRNA-seq data. Doublets/multiplets are a type of artifact that occurs when two or more cells are tagged by the same barcode, and therefore they appear as a single cell. Because this introduces non-existent transcriptional profiles, doublets can bias and mislead downstream analysis. To address this limitation computational methods to annotate and remove doublets form scRNA-seq datasets are needed.
Researchers at the University of Pittsburgh have developed vaeda, a new approach for computational annotation of doublets in scRNA-seq data. Vaeda integrates a variational auto-encoder and Positive-Unlabeled learning to produce doublet scores and binary doublet calls. The researchers apply vaeda, along with seven existing doublet annotation methods, to sixteen benchmark datasets and find that vaeda performs competitively in terms of doublet scores and doublet calls. Notably, vaeda outperforms other python-based methods for doublet annotation. All together, vaeda is a robust and competitive method for scRNA-seq doublet annotation and may be of particular interest in the context of python-based workflows.
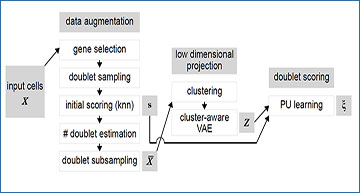
Summary of the vaeda method
Input cells X are subjected to data augmentation, where artificial doublets are simulated, a preliminary doublet score s is derived, and an augmented dataset ¯X is created. Next, a low-dimensional representationZ of ¯X is derived, using a cluster-aware variational autoencoder. Finally, positive unlabeled learning is used to derive final doublet scores ξ for each input cell/barcode.
Availability – Vaeda is available at https://github.com/kostkalab/vaeda
Schriever H, Kostka D. (2022) Vaeda computationally annotates doublets in single-cell RNA sequencing data. Bioinformatics [Epub ahead of print]. [abstract]