In the quest to understand the web of life, scientists have turned to a powerful tool called single-cell RNA sequencing (scRNA-seq). This technology allows researchers to study gene expression at the level of individual cells, providing a detailed picture of cellular functions and features. But what if we could take this a step further and compare these cellular details across different species? Such comparisons could reveal fascinating insights into how cell functions and features have evolved over time.
The Power of Single-Cell RNA Sequencing
First, let’s briefly understand what scRNA-seq is. Single-cell RNA sequencing is a method that captures the gene expression profiles of individual cells. Unlike traditional RNA sequencing, which provides an average gene expression profile across many cells, scRNA-seq reveals the unique expression pattern of each cell, uncovering the diversity and complexity within a tissue or organism.
Why Compare Across Species?
Comparing scRNA-seq data across different species can provide valuable insights into the evolutionary history of cell types and functions. By examining how gene expression patterns vary between species, researchers can trace the lineage of specific cell types and identify which genes have been conserved or changed over time. This can help us understand how certain cellular features and functions have evolved, and why they might differ between species.
The Phylogenetic Approach
Phylogenetics is the study of evolutionary relationships among biological entities—often species, genes, or cells. These relationships are typically depicted as phylogenetic trees, which are diagrams that show how different entities are related through evolutionary history.
In the context of scRNA-seq data, a phylogenetic approach involves creating trees that depict the relationships between:
- Species: How different species are related to each other.
- Genes: How different genes have evolved and diversified.
- Cells: How different cell types have emerged and evolved within and across species.
Challenges and Solutions
Comparing scRNA-seq data across species is not without its challenges. Here are some of the key hurdles and how a phylogenetic approach can help overcome them:
- Data Alignment: Aligning gene expression data from different species can be tricky due to differences in genome structure and gene annotation. Phylogenetic methods can help by aligning data based on evolutionary relationships, rather than purely sequence similarity.
- Dimensionality: scRNA-seq data is high-dimensional, meaning each cell’s gene expression profile is represented by thousands of data points. This complexity makes direct comparisons difficult. Phylogenetic methods can simplify these comparisons by focusing on evolutionary conserved features.
- Evolutionary Inference: Determining how cell functions and features have evolved requires robust statistical methods. Phylogenetic comparative approaches provide a framework for testing hypotheses about gene and cell evolution by integrating information about species relationships and gene functions.
Integrating Phylogenetics with scRNA-seq
By incorporating phylogenetic approaches into scRNA-seq analyses, researchers at Yale University have created a more comprehensive picture of cellular evolution. This integration allows for:
- Reconciliation of Different Trees: Understanding how species trees (relationships between species), gene trees (relationships between genes), and cell phylogenies (relationships between cell types) intersect and inform each other.
- Robust Hypothesis Testing: Phylogenetic methods enable researchers to test specific hypotheses about how and why certain cell functions have evolved, based on evolutionary relationships and gene expression patterns.
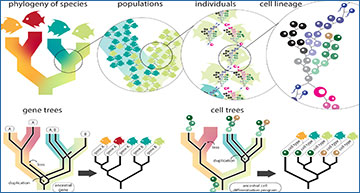
Unification of trees for species, populations, genes, and cells
All cellular life is related by an unbroken chain of cell divisions. Species phylogenies describe the relationship between populations. Populations are themselves a description of the genealogical relationships between individuals. Peering even closer reveals that each individual consists of a lineage of cells, connected to other individuals via reproductive cells. Therefore, species trees, genealogies, and cell lineages are all descriptions of the same concept—the tree of life—but at different scales. Gene trees and cell trees (i.e., cellular phylogenies) describe the evolutionary histories of specific characters within the tree of life. These trees may be discordant with species trees due to duplication, loss, and incomplete sorting in populations.
Conclusion
Comparing single-cell RNA sequencing data across species using a phylogenetic approach opens up new avenues for understanding the evolution of cellular life. By addressing the challenges of data alignment, dimensionality, and evolutionary inference, this approach provides a robust framework for exploring the relationships between genes, cells, and species. As we continue to refine these methods, we can look forward to uncovering deeper insights into the evolutionary history of life at the cellular level, paving the way for advances in biology, medicine, and beyond.
Church SH, Mah JL, Dunn CW (2024) Integrating phylogenies into single-cell RNA sequencing analysis allows comparisons across species, genes, and cells. PLoS Biol 22(5): e3002633. [article]