Lab-grown skeletal muscle cells transplanted into mice with a muscular dystrophy for 90 days are overlayed with regions of interest for spatial RNA sequencing technology used to identify how human stem cells and myofibers are communicating.
University of California, Irvine researchers have identified a gene expressed during regeneration that is critical for muscle repair. The key human skeletal muscle gene was also found in a subset of muscle fibers that were able to support human muscle stem cells after transplantation.
Although skeletal muscle is one of the most regenerative organ systems, there exists a need to improve regeneration for the more than 400 chronic muscle disorders and injuries that present clinically, including rotator cuff injuries and certain muscle disorders like Duchenne Muscular Dystrophy (DMD) or congenital muscular dystrophy.
Michael H. Hicks, PhD, assistant professor in the Department of Physiology & Biophysics at UCI School of Medicine, is the co-corresponding author of the study, along with April D. Pyle, PhD, professor in the Department of Microbiology, Immunology and Molecular Genetics at UCLA.
“With our discovery, the development of “muscle in a dish” is one step closer to reality,” said Hicks. “We’ve been researching this for years, and it’s implication for treating disease and muscle disorders and tears are immense.”
When confronted with an injury, our muscles naturally do a good job at repairing themselves. However, in severe injuries and genetic muscular diseases the muscle is unable to meet the demands of regenerating new tissues. Researchers say one solution is to take the cells from a dish and replicate how a healthy human body repairs muscle as newly generated muscle made in a lab can support stem cells better than the exhausted muscle tissue.
One disorder the Hicks lab hopes to treat with their lab-grown muscle progenitors is tears to the rotator cuff muscles, which affects up to 30% of people over the age of 65.
Damage to the rotator cuff muscles and tendon result in loss of mobility, prolonged hospitalization, and increased dependency on health care providers. Even after surgical attachment of the rotator cuff tendon to the bone, the muscle often fails to regenerate or incompletely regenerates, leading to decreased function.
Hicks was also recently funded by the UCI Anti-Cancer Challenge to use his approach for muscle reconstruction after radiation therapy for cancer survivors.
“Muscle stem cells are exposed to significant doses of radiation during radiotherapeutic management of cancer,” said Hicks. “The use of ionizing radiation has the potential to damage muscle stem cells and limit the recovery of muscle mass following disuse or overtime with age.”
Muscle stem cells are supported within anatomically defined specialized compartments, termed niches, that regulate their balance of self-renewal and differentiation over a person’s lifetime. The ability to establish new stem cells niches is essential for long-term cell therapies, in which transplanted muscle stem cells must balance the formation of new muscle fibers and maintain the stem cell pool to respond to future injuries.
Researchers demonstrated the formation of regenerating human myofibers following transplantation are a key source of niche emergence from transplanted human cells, which has previously been overlooked.
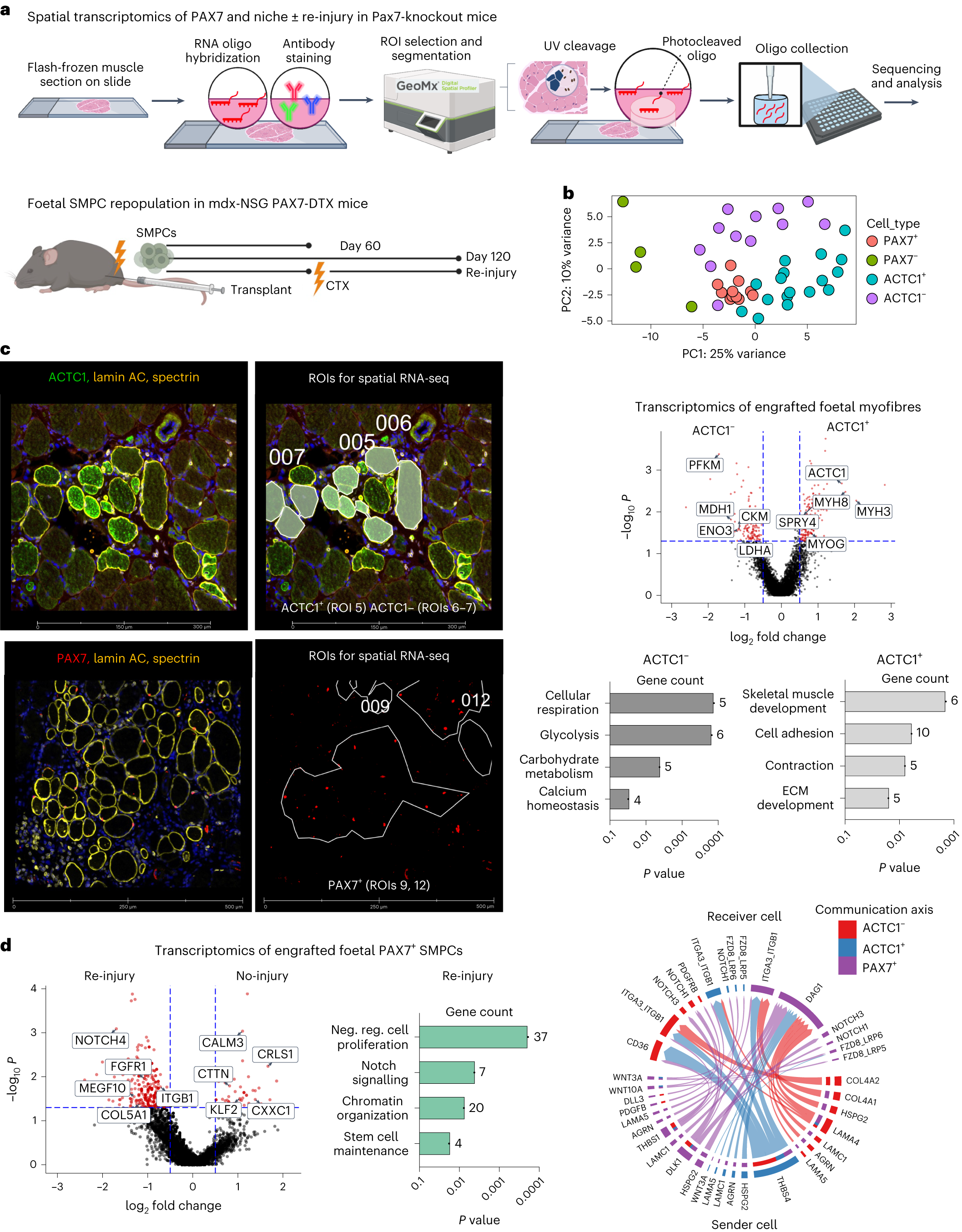
Spatial transcriptomics identifies interaction of ACTC1+ myofibres with PAX7 SMPCs in the emerging niche
a, Cartoon workflow of spatial transcriptomics on engrafted human/mouse tissue. Photocleavable RNA oligo probes are collected on ROIs for sequencing. Tissues used for spatial RNA-seq included foetal SMPCs engrafted in PAX7-ablated (or DTX) mdx-NSG mice for 60 or 120 days, and re-injury with CTX at 60 days. b, PCA plot showing clustering of engrafted human cells (PAX7+, PAX7−) and myofibres (ACTC1+, ACTC1−). Each dot represents a sequenced ROI (N = 43). c, Top left: images taken on GeoMX of day 60 engrafted foetal SMPCs show ACTC1+ and ACTC1− myofibres. GeoMx applied masks to ROIs used for sequencing. Right: volcano plot of ACTC1+ and ACTC1− myofibres. Red dots represent differential expressed genes, and selected key genes are shown. For all volcano plots differential gene expression (DGE) between ROIs was determined using GeoMx linear mixed model (LMM) statistical tests with Benjamini–Hochberg (BH) correction P < 0.05. Bar graphs show Gene Ontology of selected pathways upregulated by ACTC1+ and ACTC1− myofibres. Bottom left: PAX7 in the re-injury group and regions used for selecting single PAX7 cells for sequencing. d, Volcano plot of human foetal PAX7+ SMPCs after re-injury or no-injury 120 days after engraftment. Red dots represent differentially expressed genes, and selected key genes are shown. Graphs show Gene Ontology of foetal PAX7+ SMPCs undergoing repopulation after re-injury of selected pathways related to the SC niche comparisons between two groups. For comparing different cell types or treatments to generate volcano plot data, LMM statistical tests with BH correction were performed. Neg. reg., negative regulation. Right: CellChat analysis show predicted sender (bottom) and receiver (top) ligand and receptor interactions between foetal PAX7+ SMPCs and ACTC1+ and ACTC1− myofibres from spatial RNA-seq of engrafted tissues.
“This subset of regenerating muscle fibers resulted in a 50-fold better ability to support transplanted muscle progenitor cells,” said Pyle. “It would be interesting to determine whether myofibers in homeostasis or in disease settings could be stimulated to make skeletal muscle more regenerative and less susceptible to broad range of diseases.”
The researchers further characterized the interaction of transplanted muscle progenitor cells with the subset of muscle fibers a unique gene called ACTC1 using a new technology called spatial RNA sequencing. The equipment recently obtained by the UCI Genomics Research and Technology Hub, has a powerful ability to perform segmentation of cell types directly adjacent to one another and to obtain RNA information from those cell types.
“We tailored a high-dimensional spatial analysis platform to identify how transplanted human progenitor cells and myofibers in a mouse were communicating,” said Ben Clock, a UCI graduate student also involved in the study.
In the future, the team plans to dive deeper into restored muscle function including assessing the ability of the newly formed human muscles to connect with the motor neurons to restore motor control to the transplanted cells.
The Hicks lab at the UCI School of Medicine is pursuing both basic and translational avenues as their next steps. The ability to generate these muscle stem cells in the lab is currently under patent review by the US, Europe, and Japan. Hicks and Pyle also have plans to start a company to translate muscle stem cells for patients.
In a previous study from 2017, Hicks and Pyle made strides to create and repair skeletal muscle, termed progenitor cells, in the lab with gene editing. Yet to date, retention of human muscle progenitors after transplantation from cells grown in the lab has proven challenging.
Results from this study have identified several key receptors and ligand candidates on the muscle progenitor cells that could allow for them to interact with the myofibers, but these candidates will need to be validated before they can be used as therapeutic targets to improve muscle regeneration.
Source – UC Irvine School of Medicine
Hicks MR, Saleh KK, Clock B, Gibbs DE, Yang M, Younesi S, Gane L, Gutierrez-Garcia V, Xi H, Pyle AD. (2023) Regenerating human skeletal muscle forms an emerging niche in vivo to support PAX7 cells. Nat Cell Biol [Epub ahead of print]. [article]