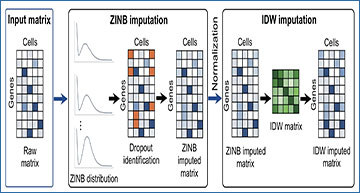
Single-cell RNA sequencing (scRNA-seq) technology has enabled discovering gene expression patterns at single cell resolution. However, due to technical limitations, there are usually excessive zeros, called ‘dropouts’, in scRNA-seq data, which may mislead the downstream analysis. Therefore, it is crucial to impute these dropouts to recover the biological information.
Researchers at Yunnan University propose a two-step imputation method called tsImpute to impute scRNA-seq data. At the first step, tsImpute adpots zero-inflated negative binomial distribution to discriminate dropouts from true zeros and performs initial imputation by calculating the expected expression level. At the second step, it conducts clustering with this modified expression matrix, based on which the final distance weighted imputation is performed. Numerical results based on both simulated and real data show that tsImpute achieves favorable performance in terms of gene expression recovery, cell clustering and differential expression analysis.
Availability: The R package of tsImpute is available at https://github.com/ZhengWeihuaYNU/tsImpute.
Zheng W, Min W, Wang S. (2023) TsImpute: An accurate two-step imputation method for single-cell RNA-seq data. Bioinformatics [Epub ahead of print]. [article]