The cellular and molecular heterogeneity of tumors is a major obstacle to cancer immunotherapy. Here, researchers at the University of Innsbruck use a systems biology approach to derive a signature of the main sources of heterogeneity in the tumor microenvironment (TME) from lung cancer transcriptomic data. The researchers demonstrate that this signature, which they called iHet, is conserved in different cancers and associated with antitumor immunity. Through the analysis of single-cell and spatial transcriptomics data, they trace back the cellular origin of the variability that explains the iHet signature. Finally, the researchers demonstrate that iHet has predictive value for cancer immunotherapy, which can be further improved by disentangling three major determinants of anticancer immune responses: activity of immune cells, immune infiltration or exclusion, and cancer-cell foreignness. This work shows how transcriptomics data can be integrated to derive a holistic representation of the phenotypic heterogeneity of the TME, and ultimately to determine its unfolding and fate during immunotherapy with immune checkpoint blockers.
Cellular origin of iHet unveiled by single-cell and spatial transcriptomics data analysis
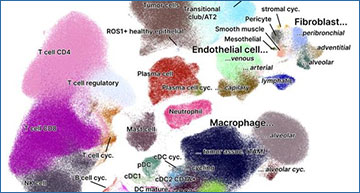
(A) UMAP plot of the full NSCLC single-cell atlas, coloured by cell-type. (B) Mean activity scores per cell type for the top transcription factors (TF, left panel) and pathways (left panel). Scores are clipped at -2.5 and 2.5, respectively. (C) Exemplary spatial transcriptomics analysis of a lung cancer slide profiled with the 10x Visium technology. The first three panels show the estimated cell-type fractions per spot for myeloid cells, fibroblasts, and CD8+ T cells, respectively. The last three panels display the log-ratio of the estimated peribronchial fibroblast vs. classical monocyte cell fractions, the TGFβ vs. TNFα pathway scores, and the TGFβ vs. NF-κB pathway scores, respectively.
Lapuente-Santana Ó, Sturm G, Kant J, Ausserhofer M, Zackl C, Zopoglou M, McGranahan N, Rieder D, Trajanoski Z, Filipe da Cunha Carvalho de Miranda N, Eduati F, Finotello F. (2023) Multimodal analysis unveils tumor microenvironment heterogeneity linked to immune activity and evasion. bioRXiv [online preprint]. [article]