Skeletal muscle fiber type distribution has implications for human health, muscle function, and performance. This knowledge has been gathered using labor-intensive and costly methodology that limited these studies. A team led by researchers at Lund University have developed a method based on muscle tissue RNA sequencing data (totRNAseq) to estimate the distribution of skeletal muscle fiber types from frozen human samples, allowing for a larger number of individuals to be tested.
By using single-nuclei RNA sequencing (snRNAseq) data as a reference, cluster expression signatures were produced by averaging gene expression of cluster gene markers and then applying these to totRNAseq data and inferring muscle fiber nuclei type via linear matrix decomposition. This estimate was then compared with fiber type distribution measured by ATPase staining or myosin heavy chain protein isoform distribution of 62 muscle samples in two independent cohorts (n = 39 and 22).
The correlation between the sequencing-based method and the other two were rATPas = 0.44 [0.13-0.67], [95% CI], and rmyosin = 0.83 [0.61-0.93], with p = 5.70 × 10-3 and 2.00 × 10-6, respectively. The deconvolution inference of fiber type composition was accurate even for very low totRNAseq sequencing depths, i.e., down to an average of ~ 10,000 paired-end reads.
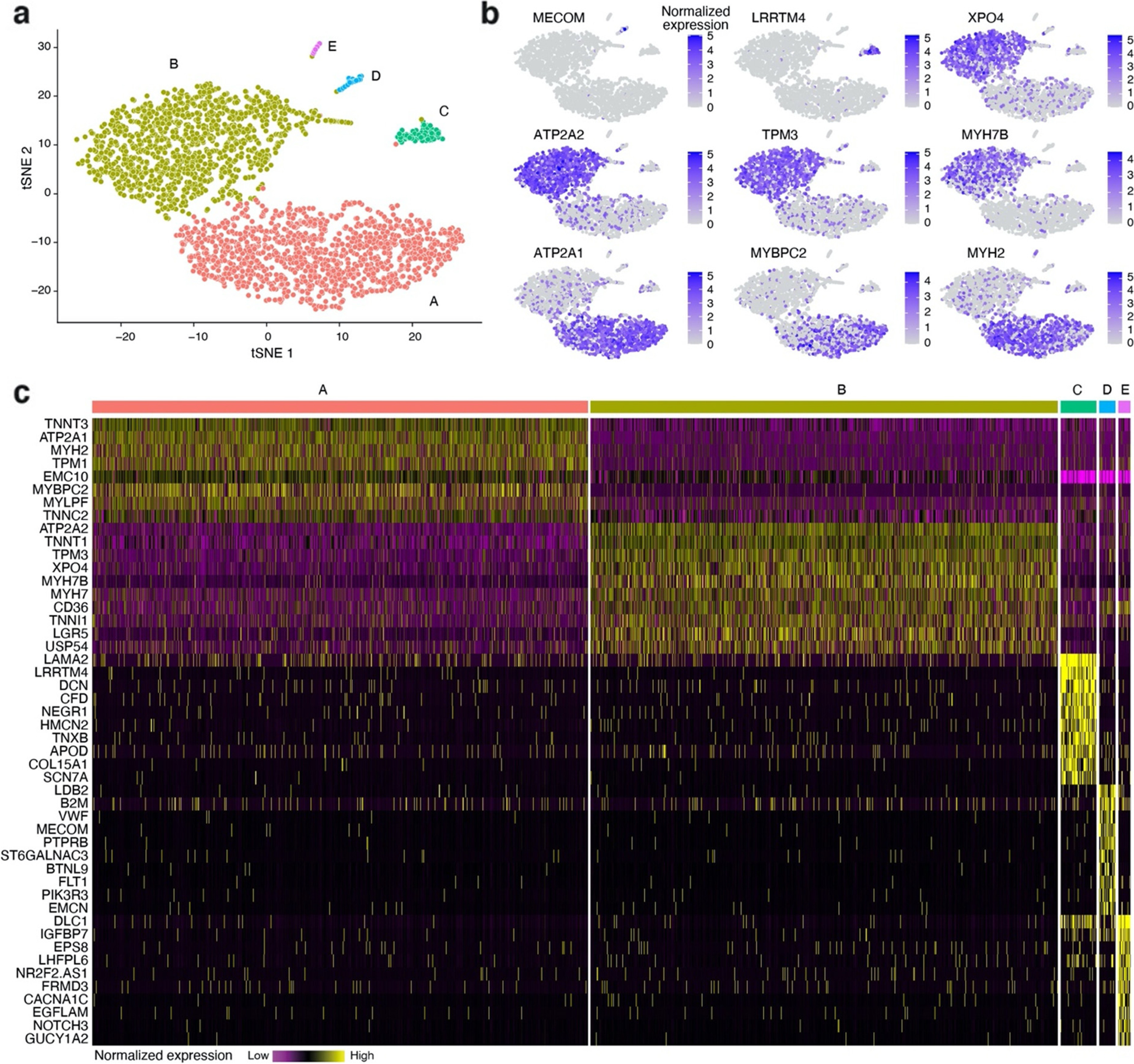
A single-nuclei RNAseq of the human skeletal muscle
Slow- (type I) and fast-twitch (type II) fibers form species-specific distinct clusters of nuclei. a Five major clusters of nuclei were identified using graph-based clustering built on Louvain modularity optimization. For visualization of the nuclei populations, t-distributed stochastic neighbor embedding (tSNE) non-linear dimension reduction was applied. b Examples of nuclei expression patterns for genes separating different clusters, i.e., ATP2A1, MYBPC2, and MYH2 are enriched in cluster A (type II fiber), XPO4, ATP2A2, TPM3, and MYH7B are enriched in cluster B (type I fiber), LRRTM4 is enriched in cluster D (endothelial), and MECOM is enriched in cluster E. c Complete list of the 48 marker genes separating the five clusters
This new method consequently allows for measurement of fiber type distribution of a larger number of samples using totRNAseq in a cost and labor-efficient way. It is now feasible to study the association between fiber type distribution and e.g. health outcomes in large well-powered studies.
Availability – https://github.com/OlaHanssonLab/PredictFiberType
Oskolkov N, Santel M, Parikh HM, Ekström O, Camp GJ, Miyamoto-Mikami E, Ström K, Mir BA, Kryvokhyzha D, Lehtovirta M, Kobayashi H, Kakigi R, Naito H, Eriksson KF, Nystedt B, Fuku N, Treutlein B, Pääbo S, Hansson O. (2022) High-throughput muscle fiber typing from RNA sequencing data. Skelet Muscle 12(1):16. [article]