Skeletal muscle fiber type distribution has implications for human health, muscle function, and performance. This knowledge has been gathered using labor-intensive and costly methodology that limited these studies. A team led by researchers at Lund University have developed a method based on muscle tissue RNA sequencing data (totRNAseq) to estimate the distribution of skeletal muscle fiber types from frozen human samples, allowing for a larger number of individuals to be tested.
The correlation between the sequencing-based method and the other two were rATPas = 0.44 [0.13-0.67], [95% CI], and rmyosin = 0.83 [0.61-0.93], with p = 5.70 × 10-3 and 2.00 × 10-6, respectively. The deconvolution inference of fiber type composition was accurate even for very low totRNAseq sequencing depths, i.e., down to an average of ~ 10,000 paired-end reads.
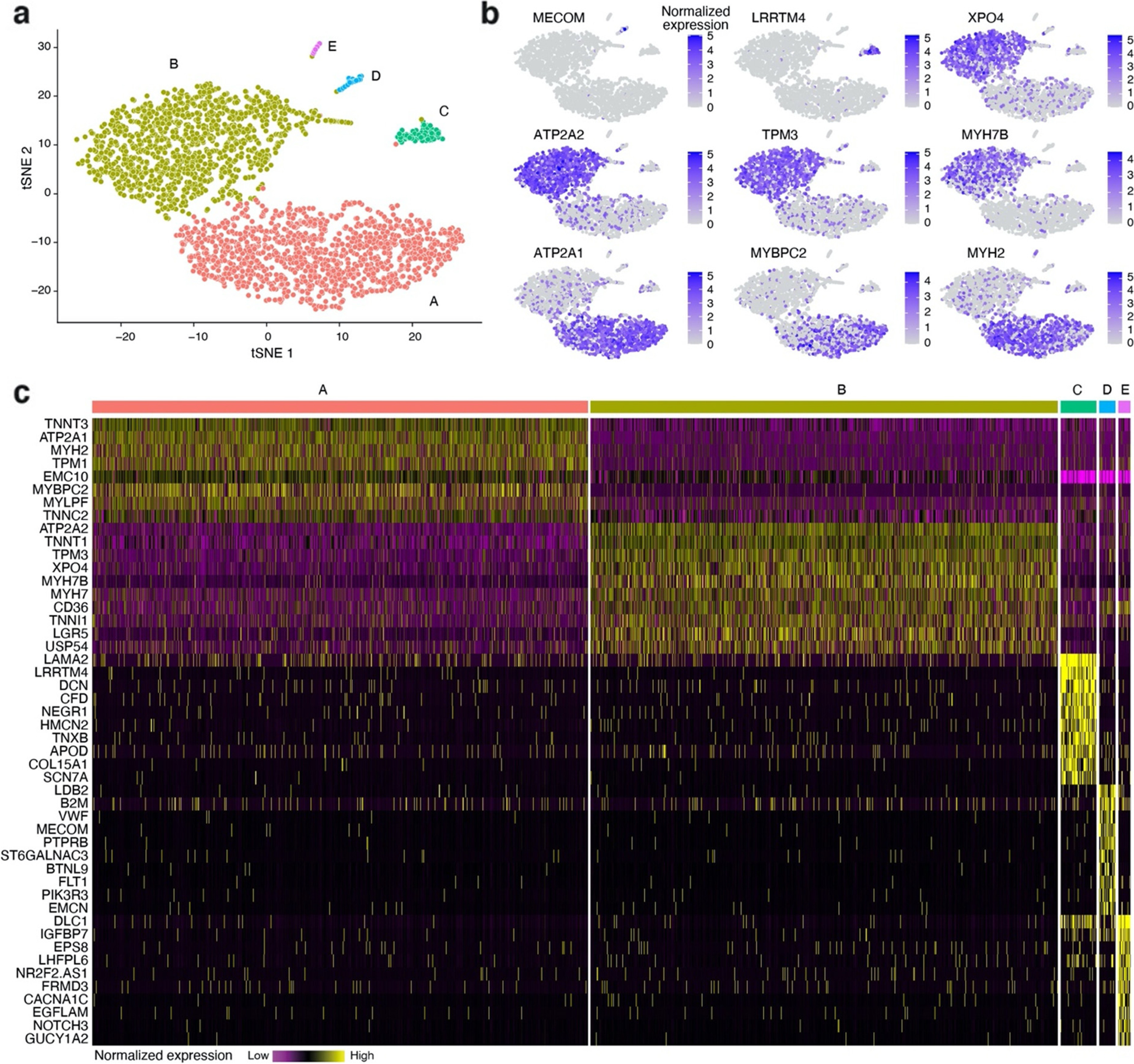
A single-nuclei RNAseq of the human skeletal muscle
Slow- (type I) and fast-twitch (type II) fibers form species-specific distinct clusters of nuclei. a Five major clusters of nuclei were identified using graph-based clustering built on Louvain modularity optimization. For visualization of the nuclei populations, t-distributed stochastic neighbor embedding (tSNE) non-linear dimension reduction was applied. b Examples of nuclei expression patterns for genes separating different clusters, i.e., ATP2A1, MYBPC2, and MYH2 are enriched in cluster A (type II fiber), XPO4, ATP2A2, TPM3, and MYH7B are enriched in cluster B (type I fiber), LRRTM4 is enriched in cluster D (endothelial), and MECOM is enriched in cluster E. c Complete list of the 48 marker genes separating the five clusters
This new method consequently allows for measurement of fiber type distribution of a larger number of samples using totRNAseq in a cost and labor-efficient way. It is now feasible to study the association between fiber type distribution and e.g. health outcomes in large well-powered studies.
Availability – https://github.com/OlaHanssonLab/PredictFiberType
Oskolkov N, Santel M, Parikh HM, Ekström O, Camp GJ, Miyamoto-Mikami E, Ström K, Mir BA, Kryvokhyzha D, Lehtovirta M, Kobayashi H, Kakigi R, Naito H, Eriksson KF, Nystedt B, Fuku N, Treutlein B, Pääbo S, Hansson O. (2022) High-throughput muscle fiber typing from RNA sequencing data. Skelet Muscle 12(1):16. [article]