Genetic regulation of gene expression is a complex process, with genetic effects known to vary across cellular contexts such as cell types and environmental conditions. A team led by researchers at the Harvard T.H. Chan School of Public Health has developed SURGE, a method for unsupervised discovery of context-specific expression quantitative trait loci (eQTLs) from single-cell transcriptomic data. This allows discovery of the contexts or cell types modulating genetic regulation without prior knowledge. Applied to peripheral blood single-cell eQTL data, SURGE contexts capture continuous representations of distinct cell types and groupings of biologically related cell types. The researchers demonstrate the disease-relevance of SURGE context-specific eQTLs using colocalization analysis and stratified LD-score regression.
SURGE model overview and simulation
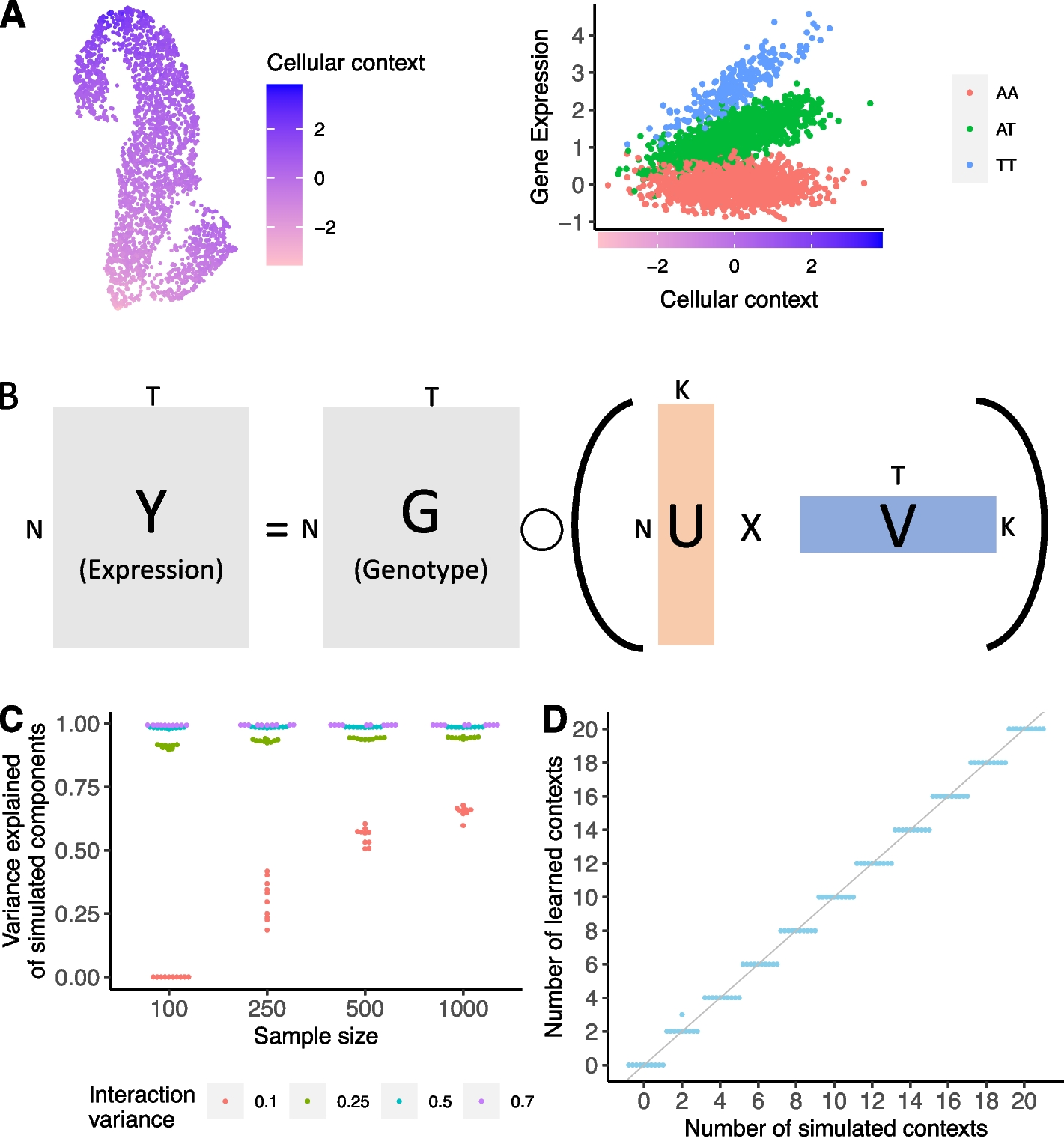
A Schematic example of an interaction eQTL where the eQTL effect size (right) changes as a function of cellular context (depicted in UMAP embedding, left). B SURGE is a novel probabilistic model that uses matrix factorization to jointly learn a continuous representation of the cellular contexts defining each measurement (U) and the corresponding eQTL effect sizes specific to each learned context (V) based on observed expression (Y) and genotype (G) data. SURGE additional accounts for the effects of known covariates and sample repeat structure on gene expression. Assume there are N samples, T genome-wide independent variant-gene pairs, and K latent contexts. C Based on simulated data, we evaluated SURGE’s ability to reconstruct simulated latent contexts as measured by the average variance explained of the simulated latent contexts by the learned latent contexts (y-axis). We simulate 5 latent contexts and vary the sample size (x-axis) and the strength of the interaction terms (colors). We fix the fraction of tests that are context-specific eQTLs for each context to .3. For each parameter setting, we run 10 independent simulations. Each dot is an independent simulation. D Based on simulated data, we evaluate SURGE’s ability to identify the number of simulated latent contexts across 10 independent simulations. The sample size was fixed to 250, the strength (variance) of the simulated interaction terms was fixed to .25, and the fraction of tests that are context-specific eQTLs for a particular context was fixed to .3. For each parameter setting, we run 10 independent simulations. Each dot is an independent simulation
Strober BJ, Tayeb K, Popp J, Qi G, Gordon MG, Perez R, Ye CJ, Battle A. (2024) SURGE: uncovering context-specific genetic-regulation of gene expression from single-cell RNA sequencing using latent-factor models. Genome Biol 25(1):28. [article]