From Alzforum by Chelsea Weidman Burke
As some scientists create huge, new, single-nucleus RNA-sequencing datasets, others devise ways to better use existing ones. At the Alzheimer’s Association International Conference held last month in San Diego, California, Evan Macosko of the Broad Institute of Harvard and MIT in Cambridge, Massachusetts, presented his work combining small databanks into a large one. Akin to doing a meta-analysis, this approach boosts statistical power, allowing Macosko to test if transcriptome changes tied to AD progression are consistent across cohorts.
Macosko and colleagues first turned to a unique cohort by Ville Leinonen, Kuopio University Hospital, Finland. Leinonen and colleagues have collected prefrontal cortex biopsies taken from 700 adults with normal-pressure hydrocephalus, a condition where excess cerebrospinal fluid swells the brain ventricles. It is treated by placing a drainage shunt into the ventricles, and this procedure allows scientists to take a tissue sample during surgery. The tissue provides a snapshot into the living brain without the peri- or postmortem artifacts that can compromise RNA stability and skew transcriptional signatures….
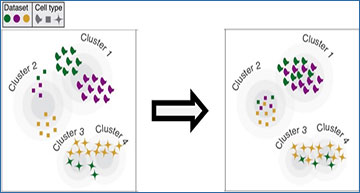
Reshuffle by Cell Type. Algorithms align cell clusters from different datasets (colors) to generate one cluster for each cell type (symbols). [Courtesy of Evan Macosko, Broad Institute.]