Wheat is a global staple food and plays a pivotal role in the livelihoods of billions of people. Although long non-coding RNAs (lncRNAs) have been recognized as crucial regulators of numerous biological processes, our knowledge of lncRNAs associated with wheat (Triticum aestivum) grain development remains minimal.
To elucidate the landscape of lncRNAs in wheat, researchers at the China Agricultural University conducted genome-wide strand-specific RNA sequencing (ssRNA-seq) on grain endosperm at 10 and 15 days after pollination (DAP) and integrated 545 publicly available transcriptome datasets from various developmental stages and tissues. The analysis pipeline was adapted from a previous study with modifications, processed RNA-seq data in three primary steps: mapping, assembly, and filtering. The results identified 20,893 lncRNAs in wheat. The characterization of these lncRNAs indicated an average transcript length of 900 bp, and were predominantly single exon structure (48%), with significant overlap with long terminal repeat retrotransposons (LTRs, 41.40%). Compared with protein-coding genes (PCGs), wheat lncRNAs exhibit shorter transcript lengths, fewer exons, and higher tissue-specific expression than PCGs, and their expression patterns were positively correlated with adjacent PCGs. Furthermore, analyzing the distribution of lncRNAs across the three wheat subgenomes (A, B, and D) revealed that 90.7% of the lncRNAs were exclusive to a single subgenome, suggesting that lncRNAs have different evolutionary trajectories compared with PCGs.
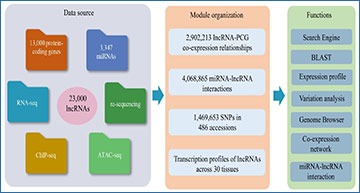
To ensure efficient access to this wealth of data, the researchers developed the comprehensive database wLNCdb, which provides various tools to explore wheat lncRNA profiles, including expression patterns, co-expression networks, functional annotations, and single nucleotide polymorphisms among wheat accessions. Notably, using wLNCdb, the researchers identified the lncRNA TraesLNC1D26001.1, which negatively regulates seed germination as its overexpression delayed wheat seed germination by upregulating Abscisic acid-insensitive 5 (TaABI5). Moreover, this lncRNA appears to co-express with genes associated with starch and protein biosynthesis in wheat, emphasizing its potential regulatory role in grain development and end-use quality.
Overview of wLNCdb and its application in exploring lncRNAs
(a) The framework of wLNCdb, including multi-omics datasets as a resource database for construction (left panel), module organization (middle panel), and interactive analysis and visualization tools (right panel). (b) A case study showing how wLNCdb can be used to explore functional lncRNAs. The ‘Search Engine’ module used a list of seed storage protein (SSP) genes as the input and identified SSP-associated lncRNAs. The ‘Expression profile’ module shows the normalized expression levels (Z-score) and tissue-specificity scores of the identified lncRNAs. For each gene or lncRNA, the tissue-specificity score was calculated by dividing the average TPM in a tissue by the sum of the average TPM values of all tissues. The ‘Co-expression network’ module shows protein-coding genes (blue nodes) co-expressed with TraesLNC1B3200 (orange node), together with their regulatory relationships (edges) acquired from wGRN[38]. The ‘miRNA-lncRNA interaction’ module shows the predicted interactions between miR168e and TraesLNC1B3200. The ‘Variation analysis’ module shows the variation profiles and haplotype distribution of TraesLNC1B3200 across 125 Chinese wheat accessions. (c) Proposed model of the role of TraesLNC1B3200 in regulating grain development and end-use quality, supported by wLNCdb.
In summation, this pioneering study provides a comprehensive map of wheat lncRNAs. The wLNCdb, with its plethora of information and advanced toolset, lays the groundwork for future exploration and analysis of the functions of lncRNAs. This research not only deepens our understanding of wheat lncRNAs’ roles, especially during seed development, but also paves the way for leveraging this knowledge to boost wheat yields and quality in the future.
Source – Eurekalert
Availability – The wLNCdb is available at: http://wheat.cau.edu.cn/wLNCdb
Zhang Z, Zhang R, Meng F, Chen Y, Wang W, et al. (2023) A comprehensive atlas of long non-coding RNAs provides insight into grain development in wheat. Seed Biology [Epub ahead of print]. [article]