The short read-length of next-generation sequencing makes it challenging to characterize highly repetitive regions (HRRs) such as centromeres, telomeres and ribosomal DNAs. Based on recent strategies that combined long-read sequencing and exogenous enzymatic labelling of open chromatin, researchers at the Southern University of Science and Technology have developed single-molecule targeted accessibility and methylation sequencing (STAM-seq) in plants by further integrating nanopore adaptive sampling to investigate the HRRs in wild-type Arabidopsis and DNA methylation mutants that are defective in CG- or non-CG methylation. The researchers found that CEN180 repeats show higher chromatin accessibility and lower DNA methylation on their forward strand, individual rDNA units show a negative correlation between their DNA methylation and accessibility, and both accessibility and CHH methylation levels are lower at telomere compared to adjacent subtelomeric region. Moreover, DNA methylation-deficient mutants showed increased chromatin accessibility at HRRs, consistent with the role of DNA methylation in maintaining heterochromatic status in plants. STAM-seq can be applied to study accessibility and methylation of repetitive sequences across diverse plant species.
STAM-seq simultaneously profiles chromatin accessibility and DNA methylation in HRR regions
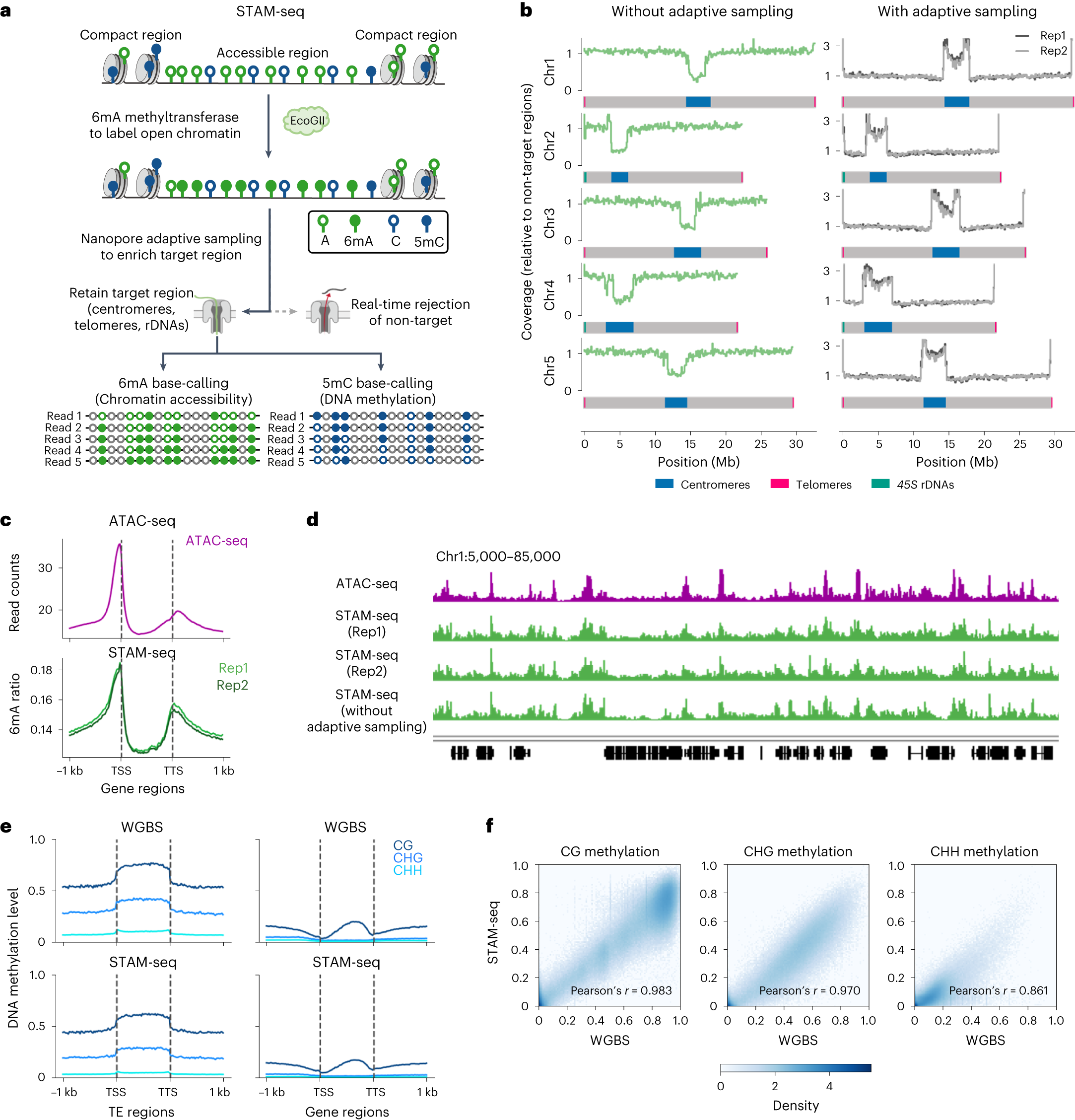
a, Outline of the STAM-seq. Open chromatin was labelled by 6mA methyltransferase. Nanopore adaptive sampling was performed to enrich sequences from HRRs during sequencing. The 6mA and 5mC modifications can be detected by base-calling within the same molecule. For 5mC base-calling, the grey circle stands for the non-C base. For 6mA base-calling, the grey circle stands for non-A base. b, Comparison of sequencing coverage in 100 kb bins of data with and without adaptive sampling. The data are normalized to the average coverage of non-target regions. The target regions include centromeres (blue), telomeres (pink) and 45S rDNA (green). c, Metaplots of chromatin accessibility signal over genes detected by ATAC-seq and STAM-seq. d, The genomic tracks in IGV show chromatin accessibility of aggregated STAM–seq data in comparison with the ATAC-seq data. e, Metaplots of DNA methylation level over transposable elements (TEs) and genes detected by WGBS and STAM-seq. f, Pairwise comparison of 5mC detection between STAM-seq and WGBS across the genome.
Mo W, Shu Y, Liu B, Long Y, Li T, Cao X, Deng X, Zhai J. (2023) Single-molecule targeted accessibility and methylation sequencing of centromeres, telomeres and rDNAs in Arabidopsis. Nat Plants [Epub ahead of print]. [abstract]