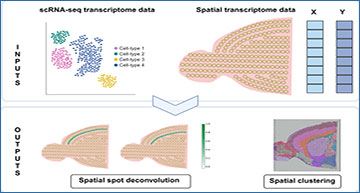
In the world of biology, understanding the details of how cells function and interact within their natural environments is crucial. One of the most advanced techniques for this purpose is spatial transcriptomics, which allows scientists to map the activity of genes within tissue samples. However, this technique comes with its own set of challenges, particularly when it comes to efficiently identifying different cell types within these samples. Enter SpatialPrompt, a new tool designed to tackle these challenges head-on.
The Challenge of Mapping Cell Types
Spatial transcriptomics involves examining gene expression in different regions of a tissue sample. However, traditional methods for analyzing this data, known as spot deconvolution tools, often fall short in two significant ways:
- Ignoring Spatial Coordinates: Many existing tools do not take into account the spatial information of the cells. This means they miss out on understanding how the physical arrangement of cells affects their function and interaction.
- Performance Issues with Large Datasets: As datasets grow in size, these tools tend to slow down significantly, making it impractical to use them for large-scale studies.
Introducing SpatialPrompt
SpatialPrompt is a game-changing tool that addresses these issues by integrating both gene expression data and spatial location information. Here’s how it works and why it’s superior:
- Integration with scRNA-seq Data: SpatialPrompt uses single-cell RNA sequencing (scRNA-seq) data as a reference to accurately determine the proportion of different cell types in spatial spots. This helps in mapping cell types more precisely within the tissue sample.
- Advanced Computational Techniques: The tool employs non-negative ridge regression and graph neural networks. These techniques allow SpatialPrompt to efficiently capture local microenvironment information, leading to more accurate results.
- Speed and Efficiency: One of the standout features of SpatialPrompt is its speed. In extensive benchmarking analyses on various datasets like Visium, Slide-seq, and MERFISH, SpatialPrompt outperformed 15 existing tools. For example, on a mouse hippocampus dataset, it could complete spot deconvolution and domain identification in just 2 minutes for 50,000 spots. This speed is 44 to 150 times faster than current methods.
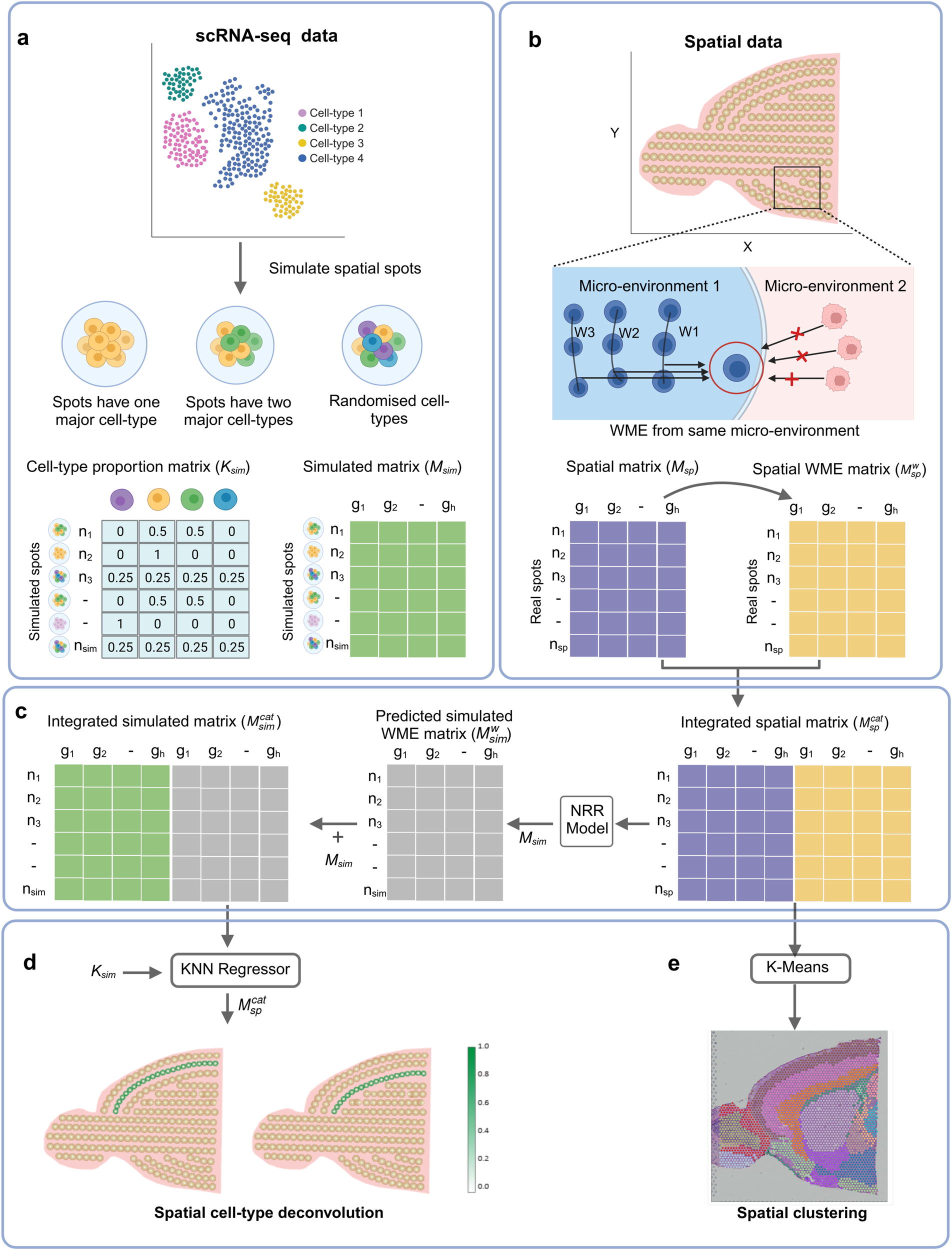
Overall workflow of SpatialPrompt
SpatialPrompt framework takes as input the spatial matrix (Msp) with spatial coordinate information and single-cell RNA-seq (scRNA-seq) matrix (Msc) with cell-type annotations. a The custom spatial spot simulator utilises Msc and cell type annotations to generate simulated expression matrix (Msim) with known cell type proportion matrix (Ksim). Spatial data is simulated under three scenarios to mimic the real spatial data, b Msp and spatial coordinates is used to calculate the weighted mean expression (WME) from neighbours in same micro-environment for each spatial spot in Msp the matrix ???? stores the WME value for nsp real spatial spots, c The non-negative ridge regression (NRR) model is built using integrated spatial matrix ??????. Next, the NRR model is employed to predict the WME for each simulated spot in Msim by utilising real spatial expression in Msp and its weighted mean neighbour expression in ????. The integrated simulated matrix ??????? is obtained by combining Msim and ????? column-wise, d For spatial deconvolution, KNN regressor model is trained on (???????) and Ksim. Then, this model predicts the cell type proportions in real spatial matrix (??????), e For domain identification, spatial clustering is performed using K-means algorithm on the integrated spatial matrix (??????). This figure created with BioRender.Com.
Superior Performance and a Vast Database
The efficiency and accuracy of SpatialPrompt are further enhanced by its integration with a curated database of over 40 scRNA-seq datasets. This seamless integration ensures that users can easily reference high-quality data for their analyses, leading to more reliable results.
Why SpatialPrompt Matters
The ability to quickly and accurately map cell types in situ has profound implications for research and medicine. For researchers, it means being able to understand the complex interactions within tissues at a much faster rate. For clinicians, it could lead to better diagnostic tools and more personalized treatments, as understanding the cellular makeup of tissues can inform on various diseases and conditions.
SpatialPrompt represents a significant advancement in the field of spatial transcriptomics. By combining cutting-edge computational techniques with a vast, integrated database, it offers unprecedented speed and accuracy in mapping cell types within tissue samples. This tool not only addresses the limitations of previous methods but also opens new avenues for research and clinical applications. As we continue to explore the complexities of cellular environments, tools like SpatialPrompt will be indispensable in our quest for deeper understanding and better healthcare solutions.
Availability – SpatiaPrompt Python package and scripts used for benchmarking analysis are available on the GitHub: https://github.com/swainasish/SpatialPrompt and at Zenodo: (https://doi.org/10.5281/zenodo.11070217).
Swain AK, Pandit V, Sharma J, Yadav P. (2024) SpatialPrompt: spatially aware scalable and accurate tool for spot deconvolution and domain identification in spatial transcriptomics. Commun Biol 7(1):639. [article]