Senescence is a state of enduring growth arrest triggered by sublethal cell damage. Given that senescent cells actively secrete proinflammatory and matrix-remodeling proteins, their accumulation in tissues of older persons has been linked to many diseases of aging. Despite intense interest in identifying robust markers of senescence, the highly heterogeneous and dynamic nature of the senescent phenotype has made this task difficult.
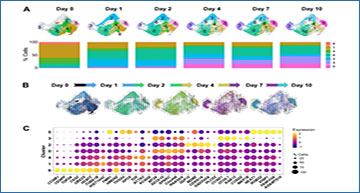
In this new study, researchers from the National Institute on Aging set out to comprehensively analyze the senescent transcriptome of human diploid fibroblasts at the individual-cell scale by performing single-cell RNA-sequencing analysis through two approaches.
“Here, we used single-cell RNA sequencing (scRNA-seq) analysis to document both the diverse transcriptomes of human senescent fibroblasts at an individual-cell scale, and the changes in the transcriptome over time during etoposide-triggered senescence.”
First, the researchers characterized the different cell states in cultures undergoing senescence triggered by different stresses, and found distinct cell subpopulations that expressed mRNAs encoding proteins with roles in growth arrest, survival and the secretory phenotype. Second, they characterized the dynamic changes in the transcriptomes of cells as they developed etoposide-induced senescence; by tracking cell transitions across this process, the researchers found two different senescence programs that developed divergently, one in which cells expressed traditional senescence markers such as p16 (CDKN2A) mRNA, and another in which cells expressed long noncoding RNAs and splicing was dysregulated. Finally, they obtained evidence that the proliferation status at the time of senescence initiation affected the path of senescence, as determined based on the expressed RNAs.
Shared and distinct transcriptomes of WI-38 fibroblasts in different senescence models
(A) WI-38 fibroblasts that were proliferating at PDL 24 (CTRL), cultured until replicative exhaustion at PDL 57 (RS), exposed to 10 Gy of ionizing radiation and cultured for an additional 10 days (IR), or cultured for six days in media containing 50 μM of etoposide and in regular media for another four days (ETO), were analyzed for SA-β-Gal activity (top), and proliferation rates evaluated by measuring by BrdU incorporation (bottom) and compared to the CTRL population. (B) Top, UMAP plots with distribution of cells that were color-coded for each group. Bottom, heatmap showing the relative expression of top marker mRNAs in each population. (C) The expression levels of representative top markers from scRNA-seq data (top) were validated by RT-qPCR analysis (bottom). The relative levels of each mRNA in CTRL, RS, IR, and ETO were first normalized to GAPDH mRNA levels and each senescence model was compared to CTRL cells. In (A, C) significance was assessed by two-tailed unpaired Student’s t-test, n = 2, *p < 0.05, **p < 0.01, ***p < 0.001.
“We propose that a deeper understanding of the transcriptomes during the progression of different senescent cell phenotypes will help develop more effective interventions directed at this detrimental cell population.”
Source – Eurekalert
Wechter N, Rossi M, Anerillas C, Tsitsipatis D, Piao Y, Fan J, Martindale JL, De S, Mazan-Mamczarz K, Gorospe M. (2023) Single-cell transcriptomic analysis uncovers diverse and dynamic senescent cell populations. Aging (Albany NY) 15(8):2824-2851. [article]