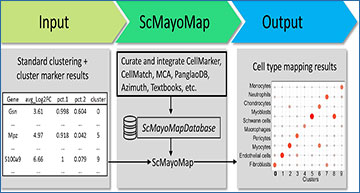
Single-cell RNA-sequencing (scRNA-seq) has become a widely used tool for both basic and translational biomedical research. In scRNA-seq data analysis, cell type annotation is an essential but challenging step. In the past few years, several annotation tools have been developed. These methods require either labeled training/reference datasets, which are not always available, or a list of predefined cell subset markers, which are subject to biases. Thus, a user-friendly and precise annotation tool is still critically needed.
Mayo Clinic researchers curated a comprehensive cell marker database named scMayoMapDatabase and developed a companion R package scMayoMap, an easy-to-use single-cell annotation tool, to provide fast and accurate cell type annotation. The effectiveness of scMayoMap was demonstrated in 48 independent scRNA-seq datasets across different platforms and tissues. Additionally, the scMayoMapDatabase can be integrated with other tools and further improve their performance.
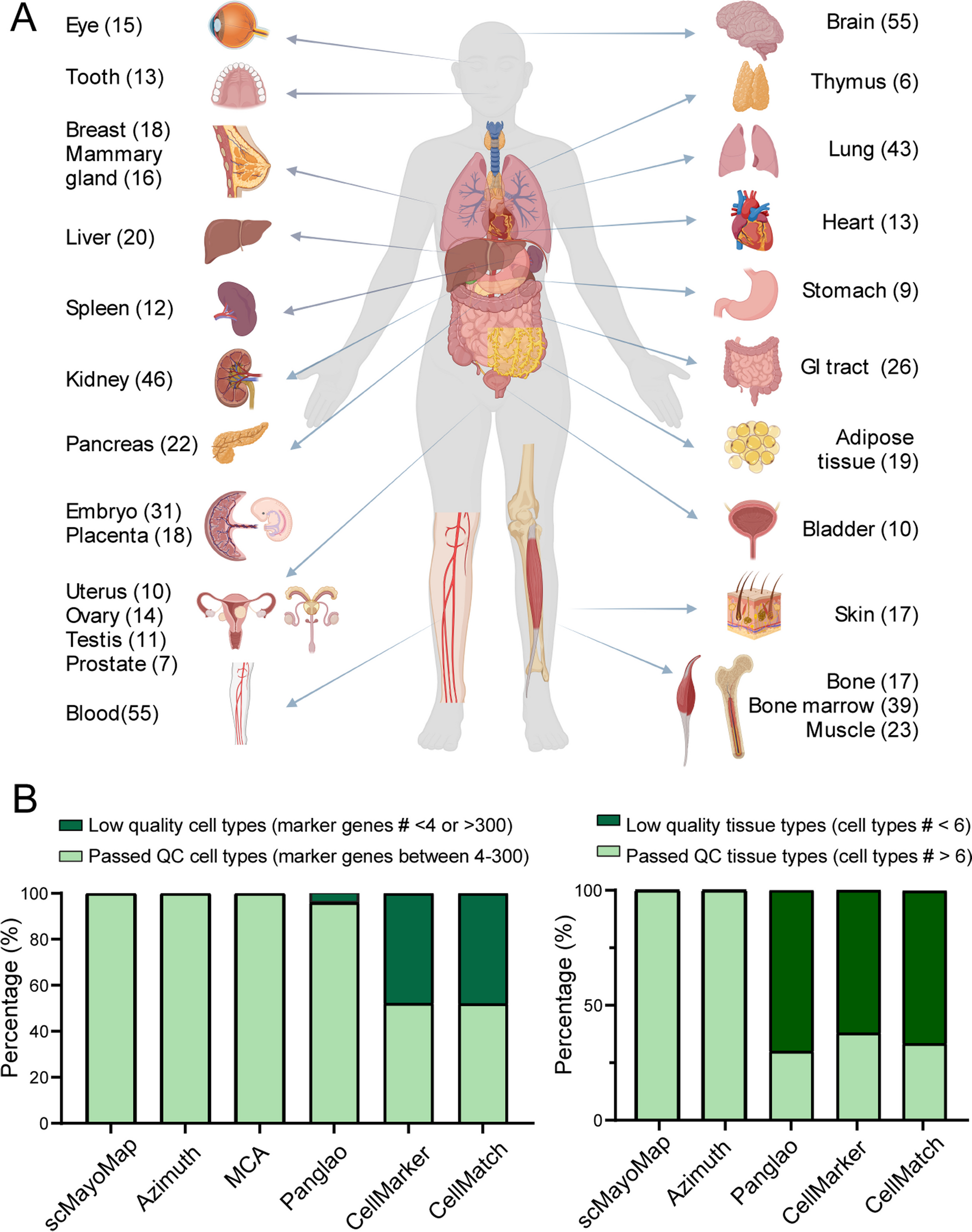
Summary of the scMayoMapDatabase
A The scMayoMapDatabase contains 28 tissues and 340 cell types for human and mouse. The number of cell types within each tissue was labeled in parentheses. B Comparison of scMayoMapDatabase to other public databases. The left panel shows the cell type information and the right panel shows the tissue information in each database
scMayoMap and scMayoMapDatabase will help investigators to define the cell types in their scRNA-seq data in a streamlined and user-friendly way.
Availability – scMayoMap is an open-source software, available at GitHub (https://github.com/chloelulu/scMayoMap)
Yang L, Ng YE, Sun H, Li Y, Chini LCS, LeBrasseur NK, Chen J, Zhang X. (2023) Single-cell Mayo Map (scMayoMap): an easy-to-use tool for cell type annotation in single-cell RNA-sequencing data analysis. BMC Biol 21(1):223. [article]