Single-cell RNA sequencing (scRNA-seq) has emerged as a vital tool in tumour research, enabling the exploration of molecular complexities at the individual cell level. It offers new technical possibilities for advancing tumour research with the potential to yield significant breakthroughs. However, deciphering meaningful insights from scRNA-seq data poses challenges, particularly in cell annotation and tumour subpopulation identification. Efficient algorithms are therefore needed to unravel the intricate biological processes of cancer. To address these challenges, benchmarking datasets are essential to validate bioinformatics methodologies for analysing single-cell omics in oncology.
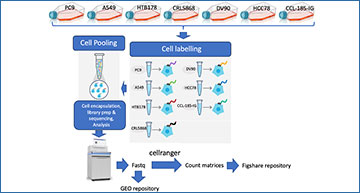
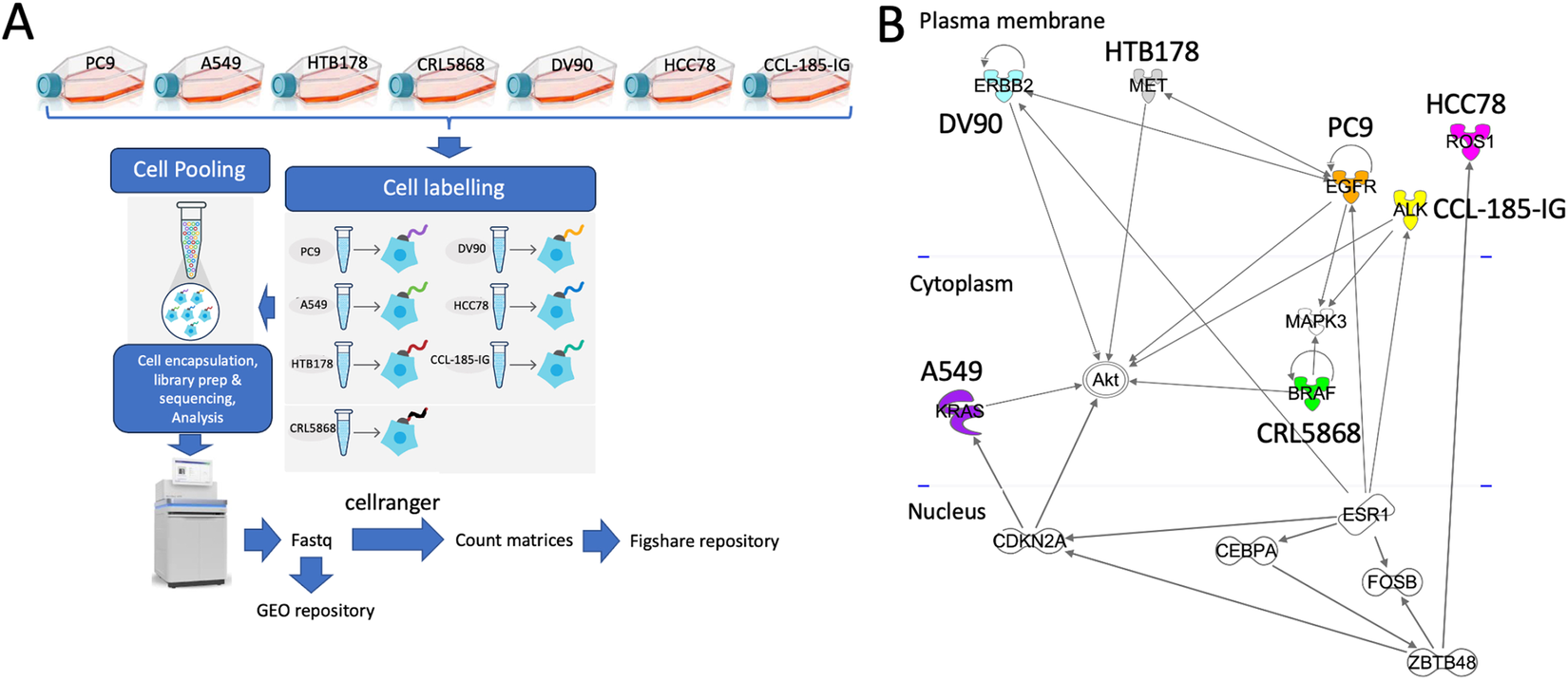
Researchers at the University of Torino present a 10XGenomics scRNA-seq experiment, providing a controlled heterogeneous environment using lung cancer cell lines characterised by the expression of seven different driver genes (EGFR, ALK, MET, ERBB2, KRAS, BRAF, ROS1), leading to partially overlapping functional pathways. This dataset provides a comprehensive framework for the development and validation of methodologies for analysing cancer heterogeneity by means of scRNA-seq.
Single cell RNAseq benchmark experiment embedding “controlled” cancer heterogeneity
(A) Outline the experimental workflow (B) Functional relationships among EGFR, ALK, MET, ERBB2, KRAS, BRAF, ROS1 cancer driver genes.
Arigoni M, Ratto ML, Riccardo F, Balmas E, Calogero L, Cordero F, Beccuti M, Calogero RA, Alessandri L. (2024) A single cell RNAseq benchmark experiment embedding “controlled” cancer heterogeneity. Sci Data 11(1):159. [article]