While recent advances in plant single-cell RNA sequencing (scRNA-seq) have made numerous strides in identifying novel regulatory events, transcriptional profiling of certain cell types, such as phloem poles, has not yet been thoroughly investigated.
Phloem poles, structures involved in long-distance transport of sugar and other metabolites, are characterized by four heterogeneous cell types: protophloem sieve elements, metaphloem sieve elements, phloem pole pericycles and companion cells. As most of these cell types play an important role in phloem pole development, they were found underrepresented in previous scRNA-seq studies. To deconvolute this heterogeneity, researchers at the University of Cambridge combined cell-type specific marker lines, fluorescence-activated cell sorting and SMRT-sequencing technologies to obtain high-resolution scRNA-seq profiles within protophloem sieve element-neighboring regions across developmental stages. The authors successfully sorted and enriched cell types based on distinct fluorescent translational fusion lines, including sieve element, companion cell and phloem pole pericycle, at a continuity of developmental stages.
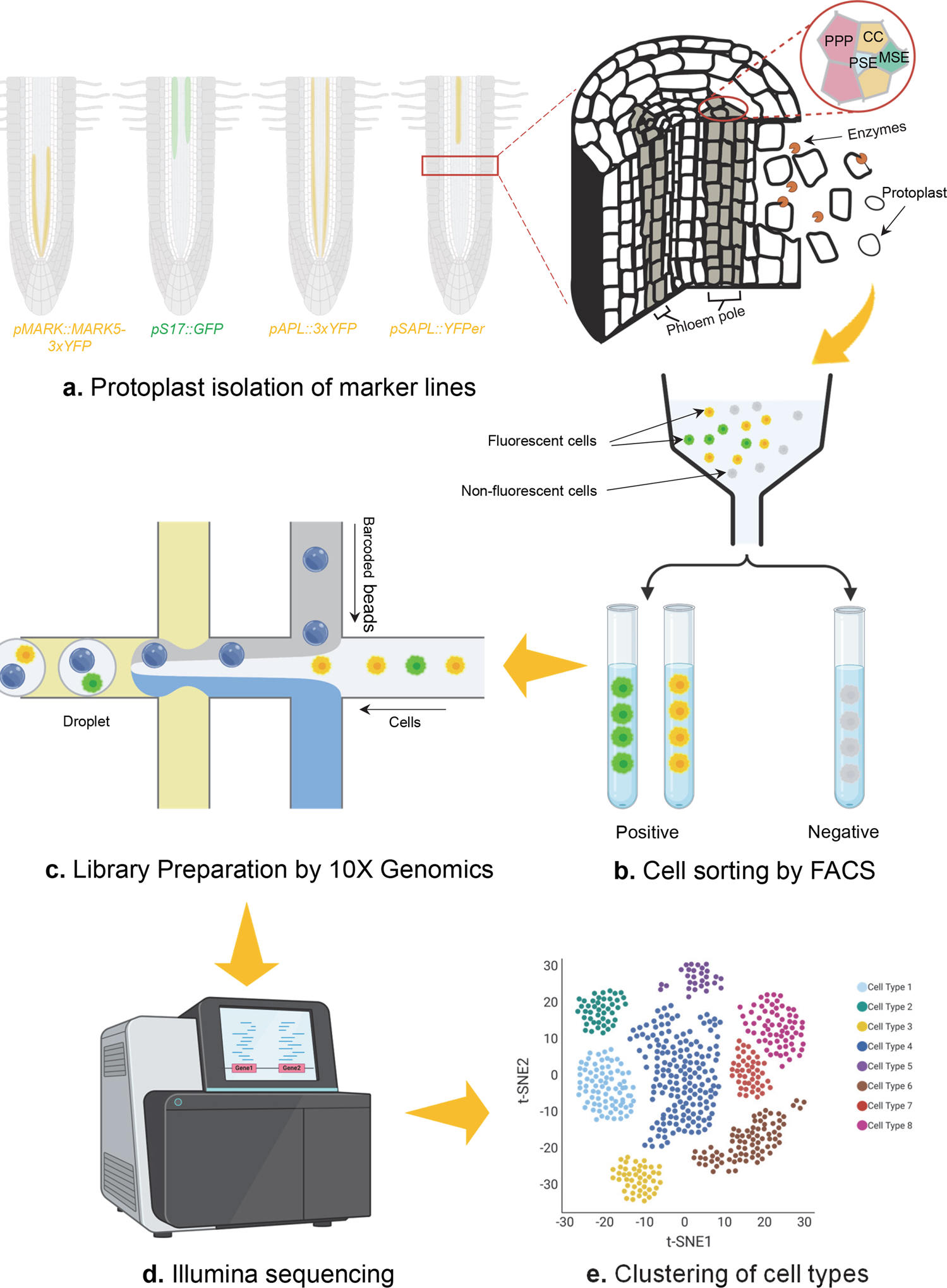
Workflow of second-generation scRNA-seq
Cells derived from marker lines specifying cell identities are dissociated via protoplast isolation (a), followed by fluorescence-activated cell sorting (FACS) to enrich target positive cells and eliminate unwanted negative cells (b). Positive cells collected are later used to prepare cDNA library within individual droplet (c) and subsequently got sequenced (d). Based on transcriptomic data coming from single cells, cell identities and development atlas are finally profiled (e).
This study provides a broad perspective of the developmental trajectories of the phloem poles as well as highlights the critical importance of DOF TFs to phloem development. This study also outlines a technical framework that can potentially be extended to other plant organs to determine spatiotemporal gene regulation both at steady state and under diverse environmental conditions.
Source – Nature Communications Biology
Liu J, Majeed A, Mukhtar MS. (2022) Rooting through single-cell sequencing in phloem pole cells. Commun Biol 5(1):1194. [article]
Otero S, Gildea I, Roszak P, Lu Y, Di Vittori V, Bourdon M, Kalmbach L, Blob B, Heo JO, Peruzzo F, Laux T, Fernie AR, Tavares H, Helariutta Y. (2022) A root phloem pole cell atlas reveals common transcriptional states in protophloem-adjacent cells. Nat Plants 8(8):954-970. [abstract]