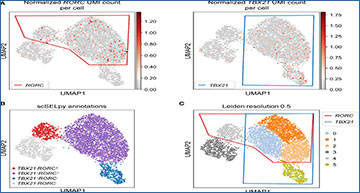
Single cell RNA sequencing plays an increasing and indispensable role in immunological research such as in the field of inflammatory bowel diseases (IBD). Professional pipelines are complex, but tools for the manual selection and further downstream analysis of single cell populations are missing so far. Researchers at Friedrich-Alexander-Universität Erlangen-Nürnberg have developed a tool called scSELpy, which can easily be integrated into Scanpy-based pipelines, allowing the manual selection of cells on single cell transcriptomic datasets by drawing polygons on various data representations. The tool further supports the downstream analysis of the selected cells and the plotting of results. Taking advantage of two previously published single cell RNA sequencing datasets, the researchers show that this tool is useful for the positive and negative selection of T cell subsets implicated in IBD beyond standard clustering. They further demonstrate the feasibility for subphenotyping T cell subsets and use scSELpy to corroborate earlier conclusions drawn from the dataset. Moreover, we also show its usefulness in the context of T cell receptor sequencing. Collectively, scSELpy is a promising additive tool fulfilling a so far unmet need in the field of single cell transcriptomic analysis that might support future immunological research.
Selection and removal of central memory T cells from the dataset
using scSELpy to obtain effector memory T cells
(A) Selection of regions enriched for cells expressing CCR7 and SELL (CD62L, L-Selectin) on UMAP plots. (B) Identification and removal of the cells located in both the CCR7 and SELL polygon as selected in A (green). (C) Re-analysis of the remaining cells from raw data. Normalization and UMAP dimension reduction is performed anew. (D) Scatter plot of all cells from the dataset with normalized CCR7 and SELL expression on the x-axis and y-axis, respectively. On this scatter plot, scSELpy was employed to categorize cells with or without expression of CCR7 and/or SELL. (E) UMAP density plots highlighting the cells belonging to the categories created in (D). The parameter “vmin” to control the lower limit in the color scale was set to 0.05.
Availability – scSELpy is available for download at https://github.com/MarkDedden/scSELpy together with installation instructions, the data analysis pipeline and a link to the documentation, which includes a tutorial.
Dedden M, Wiendl M, Müller TM, Neurath MF, Zundler S. (2023) Manual cell selection in single cell transcriptomics using scSELpy supports the analysis of immune cell subsets. Front Immunol [Epub ahead of print]. [article]