Single-cell RNA sequencing (scRNA-seq) is nowadays widely used to measure gene expression in individual cells, but meaningful biological interpretation of the generated scRNA-seq data remains a complicated task. Indeed, expertise in both the biological domain under study, statistics, and computer programming are prerequisite for thorough analysis of scRNA-seq data. However, biological experts may lack data science expertise, and bioinformaticians limited understanding of the biology may lead to time-consuming iterations. A user-friendly and automated workflow with possibility for customization is hence of a wide interest for both the biological and bioinformatics communities, and for their fruitful collaborations.
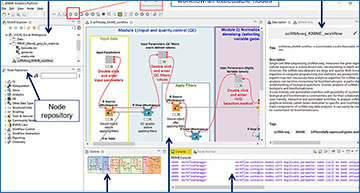
Researchers from the Aix Marseille Université have developed a locally installable, user-friendly, interactive, and automated workflow that allows the users to perform the main steps of scRNA-seq data analysis. The interface is composed of graphical entities dedicated to specific and modifiable tasks. It can easily be used by biologists and can also serve as a customizable basis for bioinformaticians.
Overview of the scRNAseq_KNIME workflow
Availability – The workflow is publicly available at https://github.com/Saminakausar/scRNAseq_KNIME.
Kausar S, Asif M, Baudot A. (2023) scRNAseq_KNIME workflow: A Customizable, Locally Executable, Interactive and Automated KNIME workflow for single-cell RNA seq. bioRXiv [online pre-print]. [abstract]