Recent developments in single-cell technology have enabled the exploration of cellular heterogeneity at an unprecedented level, providing invaluable insights into various fields, including medicine and disease research. Cell type annotation is an essential step in its omics research. The mainstream approach is to utilize well-annotated single-cell data to supervised learning for cell type annotation of new singlecell data. However, existing methods lack good generalization and robustness in cell annotation tasks, partially due to difficulties in dealing with technical differences between datasets, as well as not considering the heterogeneous associations of genes in regulatory mechanism levels.
Researchers at ShenZhen University have developed the scPML model, which utilizes various gene signaling pathway data to partition the genetic features of cells, thus characterizing different interaction maps between cells. Extensive experiments demonstrate that scPML performs better in cell type annotation and detection of unknown cell types from different species, platforms, and tissues.
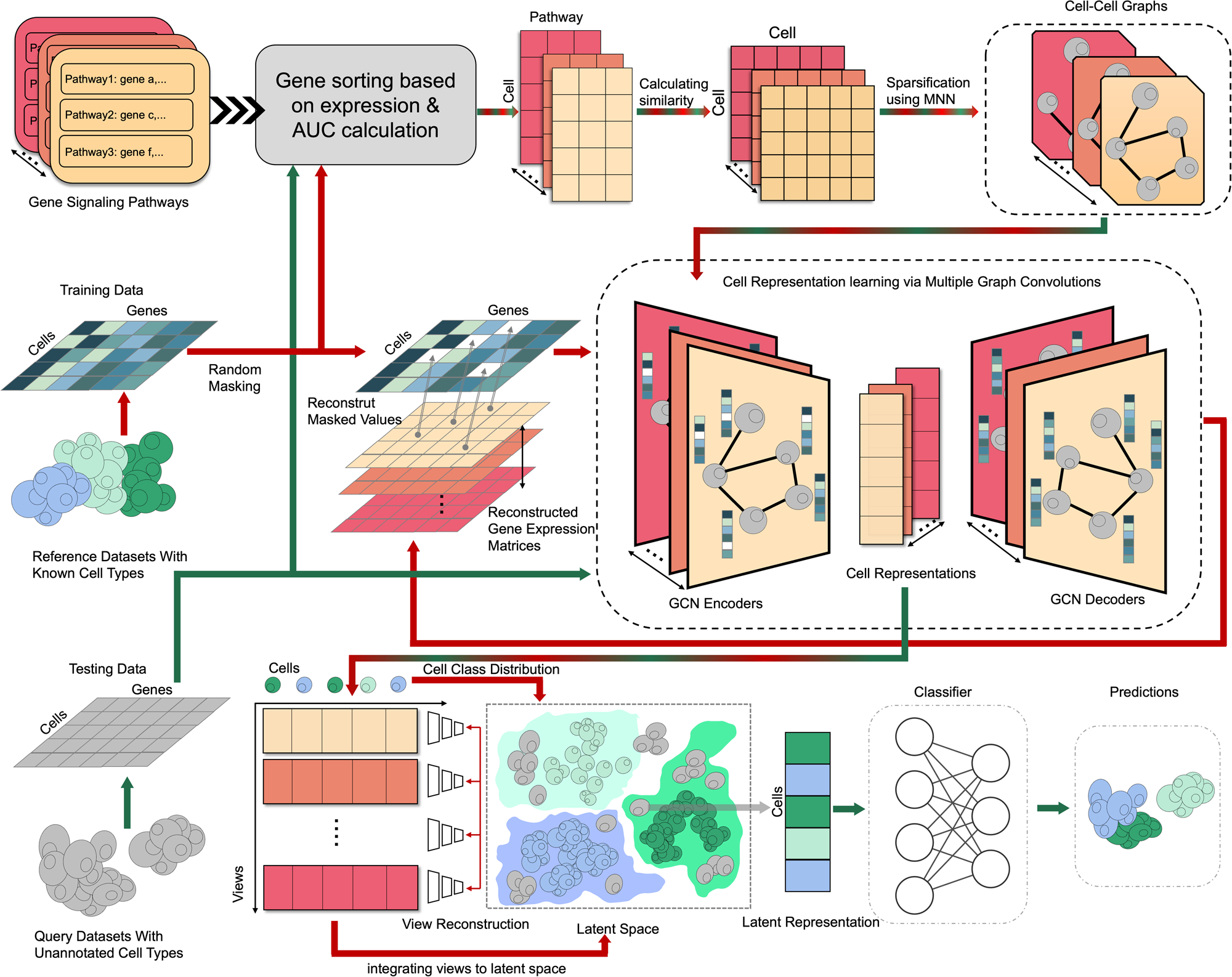
Overview of scPML
a scPML constructing cell-cell graphs using gene enrichment analysis with various pathways, yielding various cell-cell graphs marked with different colors. b Self-supervised GCN auto-encoder with the objective of recovering the masked units of processed expression data. The white grids are masked values which will be set to 0. c Obtaining common latent representation with multiple embeddings using multi-view learning. scPML attempts to find a common representation which can be reconstructed to according embeddings and has the quality of separability. After obtaining the common latent representations, scPML uses a classifier to assign labels.
Du ZH, Hu WL, Li JQ, Shang X, You ZH, Chen ZZ, Huang YA. (2023) scPML: pathway-based multi-view learning for cell type annotation from single-cell RNA-seq data. Commun Biol 6(1):1268. [article]