Scientists have long grappled with the challenge of deciphering gene expression profiles at the single-cell level, a task made possible through a cutting-edge technique known as single-cell RNA sequencing (scRNA-seq). However, while scRNA-seq offers unprecedented insights into cellular heterogeneity, it comes with its own set of hurdles, including technical noise, dropout events, and sparsity in the data.
A recent study by researchers at KAIST has sought to tackle these obstacles head-on, presenting a novel approach called single-cell bilevel feature propagation (scBFP). This two-step method harnesses the power of graph-based feature propagation to enhance the quality of scRNA-seq data and unlock hidden biological truths.
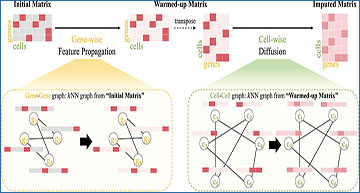
The overall framework of scBFP
Given an initial gene–cell count matrix, scBFP first conducts Gene-wise Feature Propagation on a gene–gene graph derived from this matrix, yielding a warmed-up matrix. Subsequently, using this warmed-up matrix, we derive an enhanced cell–cell graph and carry out diffusion to obtain the final imputed matrix.
First, scBFP addresses the issue of dropout events, where certain genes fail to be detected in individual cells. To combat this, the method imputes zero values (indicative of undetected genes) using non-zero values from neighboring cells. This clever strategy ensures that the imputation process doesn’t inadvertently affect the expression levels of genes that were actually detected, safeguarding the integrity of the data.
Once the zero values have been imputed, scBFP moves on to the denoising stage. Here, it capitalizes on the inherent relationships between genes and cells, leveraging graph structures to refine the dataset. Unlike previous approaches that focused solely on either gene-gene or cell-cell associations, scBFP takes a holistic approach, considering both types of relationships to achieve comprehensive denoising.
The researchers conducted extensive experimental tests on real scRNA-seq data. These experiments have demonstrated the effectiveness of scBFP across various downstream tasks, from clustering cells based on similar expression patterns to identifying key genes driving cellular diversity. In doing so, scBFP has unlocked valuable biological insights that were previously obscured by technical noise and sparsity.
In summary, the advent of scBFP represents a significant breakthrough in the field of single-cell RNA sequencing analysis. By addressing the challenges of technical noise, dropout events, and sparsity with its innovative two-step approach, scBFP has paved the way for a deeper understanding of cellular heterogeneity and opened new avenues for biological exploration.
Lee J, Yun S, Kim Y, Chen T, Kellis M, Park C. (2024) Single-cell RNA sequencing data imputation using bi-level feature propagation. Brief Bioinform 25(3):bbae209. [article]