Automatic cell type annotation methods are increasingly used in single-cell RNA sequencing (scRNA-seq) analysis due to their fast and precise advantages. However, current methods often fail to account for the imbalance of scRNA-seq datasets and ignore information from smaller populations, leading to significant biological analysis errors.
Researchers at the Chinese University of Hong Kong have developed scBalance, an integrated sparse neural network framework that incorporates adaptive weight sampling and dropout techniques for auto-annotation tasks. Using 20 scRNA-seq datasets with varying scales and degrees of imbalance, the researchers demonstrate that scBalance outperforms current methods in both intra- and inter-dataset annotation tasks. Additionally, scBalance displays impressive scalability in identifying rare cell types in million-level datasets, as shown in the bronchoalveolar cell landscape. scBalance is also significantly faster than commonly used tools and comes in a user-friendly format, making it a superior tool for scRNA-seq analysis on the Python-based platform.
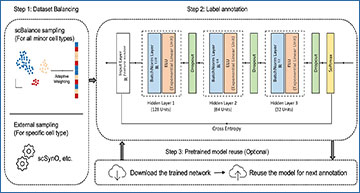
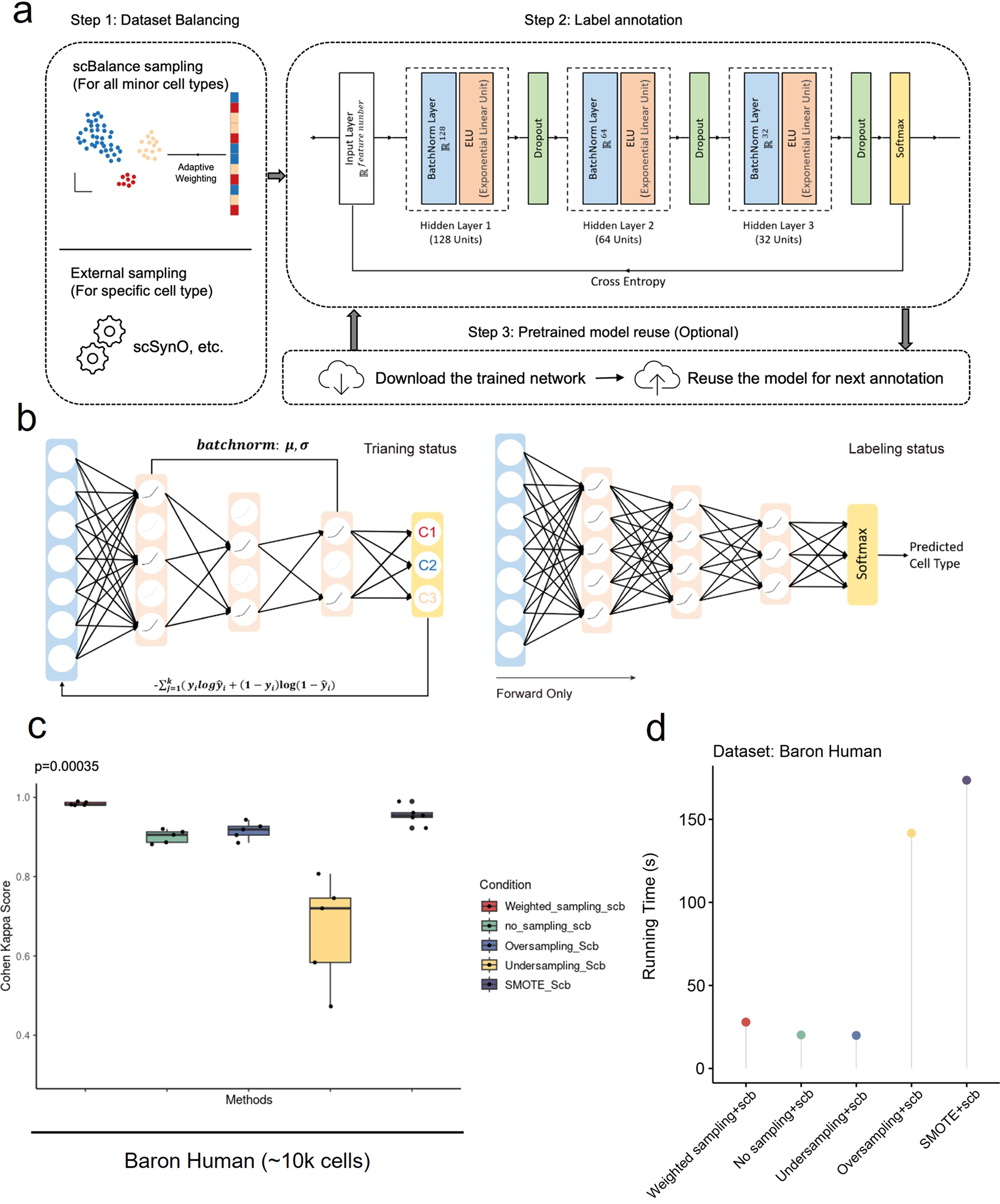
Schematic overview of scBalance
a The method is constructed based on the supervised learning framework, which contains a dataset-balancing module and a dropout neural network module. Step 1 Upper: With our adaptive weighted sampling, scBalance will automatically choose the weight for each cell type in the reference dataset and construct the training batch. Lower: Users can choose an external dataset-balancing method, such as scSynO, instead of using our internal balancing method. Only the classifier will be used in this case. Step 2: While training, scBalance will iteratively learn mini batches from a three-layer neural network until the cross-entropy loss converges. b Dropout setting in different stages. In the training stage, scBalance randomly disables neurons in the network. The dropout layer is binary with a rate of 0.5. All the dropped units will be reconnected in the testing stage. The prediction will be processed by a fully connected neural network. c Evaluation of balancing methods shows that our sampling method outperforms simple oversampling and downsampling methods as well as the SMOTE method. The p-value is from a significance test of scBalance and SMOTE (n = 5 for each boxplot). d Comparison of running times among different sampling techniques.
Availability – scBalance is available as an independent Python package at https://github.com/yuqcheng/scBalance.
Cheng Y, Fan X, Zhang J, Li Y. (2023) A scalable sparse neural network framework for rare cell type annotation of single-cell transcriptome data. Commun Biol 6(1):545. [article]