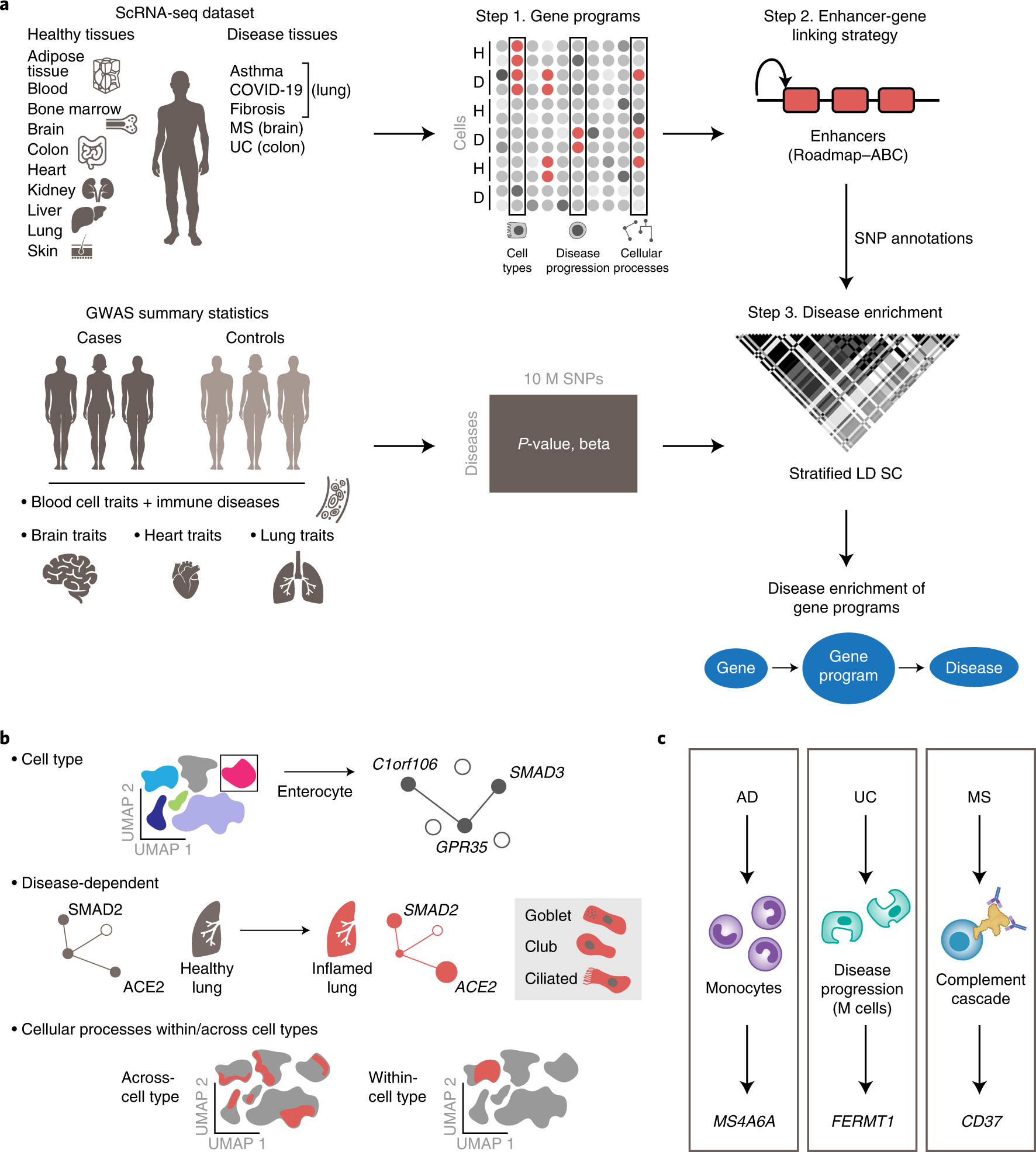
Genome-wide association studies provide a powerful means of identifying loci and genes contributing to disease, but in many cases, the related cell types/states through which genes confer disease risk remain unknown. Deciphering such relationships is important for identifying pathogenic processes and developing therapeutics. Researchers from the Broad Institute of MIT and Harvard have developed sc-linker, a framework for integrating single-cell RNA-sequencing, epigenomic SNP-to-gene maps and genome-wide association study summary statistics to infer the underlying cell types and processes by which genetic variants influence disease. The inferred disease enrichments recapitulated known biology and highlighted notable cell-disease relationships, including γ-aminobutyric acid-ergic neurons in major depressive disorder, a disease-dependent M-cell program in ulcerative colitis and a disease-specific complement cascade process in multiple sclerosis. In autoimmune disease, both healthy and disease-dependent immune cell-type programs were associated, whereas only disease-dependent epithelial cell programs were prominent, suggesting a role in disease response rather than initiation. This framework provides a powerful approach for identifying the cell types and cellular processes by which genetic variants influence disease.
Approach for identifying disease-critical cell types and cellular processes
by integration of single-cell profiles and human genetics
a, The sc-linker framework. Left: input. scRNA-seq (top) and GWAS (bottom) data. Middle and right: step 1: deriving cell-type, disease-dependent and cellular process gene programs from scRNA-seq (top) and associating SNPs with traits from human GWASs (bottom). Step 2: generation of SNP annotations. Gene programs are linked to SNPs by enhancer–gene-linking strategies to generate SNP annotations. Step 3: S-LDSC is applied to the resulting SNP annotations to evaluate heritability enrichment for a trait. b, Constructing gene programs. Top: cell-type programs of genes specifically expressed in one cell type versus others. Middle: disease-dependent programs of genes specifically expressed in cells of the same type in disease versus healthy samples. Bottom: cellular process programs of genes co-varying either within or across cell subsets; these programs may be healthy specific, disease specific or shared. c, Examples of disease–gene, program–gene relationships recovered by our framework.
Jagadeesh KA, Dey KK, Montoro DT, Mohan R, Gazal S, Engreitz JM, Xavier RJ, Price AL, Regev A. (2022) Identifying disease-critical cell types and cellular processes by integrating single-cell RNA-sequencing and human genetics. Nat Genet 54(10):1479-1492. [article]