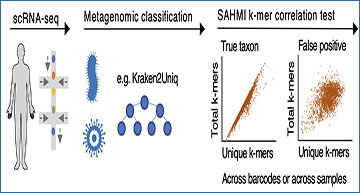
Existing genomic sequencing data can be used to study host–microbiome ecosystems; however, distinguishing signals that originate from truly present microbes from contaminating species and artifacts is a substantial and often prohibitive challenge. Researchers from Rutgers University show that emerging sequencing technologies definitely capture reads from present microbes. The researchers developed SAHMI, a computational resource to identify truly present microbial nucleic acids, as well as filter contaminants and spurious false-positive taxonomic assignments from standard transcriptomic sequencing of mammalian tissues. In benchmark studies, SAHMI correctly identifies known microbial infections present in diverse tissues, and the researchers validate SAHMI’s enrichment for correctly classified, truly present species using multiple orthogonal computational experiments. The application of SAHMI to single-cell and spatial genomic data thus enables co-detection of somatic cells and microorganisms and joint analysis of host–microbiome ecosystems.
A schematic representation of the SAHMI workflow
Availability – The SAHMI pipeline is available on our Github (https://github.com/sjdlabgroup/SAHMI) and at Zenodo (https://doi.org/10.5281/zenodo.7017103)
Ghaddar B, Blaser MJ, De S. (2023) Denoising sparse microbial signals from single-cell sequencing of mammalian host tissues. Nat Comput Sci [Epub ahead of print]. [abstract]