RNAseq data can be used to infer genetic variants, yet its use for estimating genetic population structure remains underexplored. Researchers at the Baker Heart and Diabetes Institute have developed a freely available computational tool (RGStraP) to estimate RNAseq-based genetic principal components (RG-PCs) and assess whether RG-PCs can be used to control for population structure in gene expression analyses. Using whole blood samples from understudied Nepalese populations and the Geuvadis study, the researchers show that RG-PCs had comparable results to paired array-based genotypes, with high genotype concordance and high correlations of genetic principal components, capturing subpopulations within the dataset. In differential gene expression analysis, they found that inclusion of RG-PCs as covariates reduced test statistic inflation. These data demonstrates that genetic population structure can be directly inferred and controlled for using RNAseq data, thus facilitating improved retrospective and future analyses of transcriptomic data
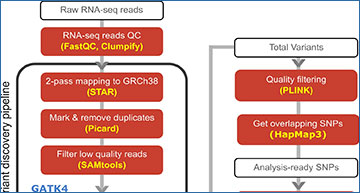
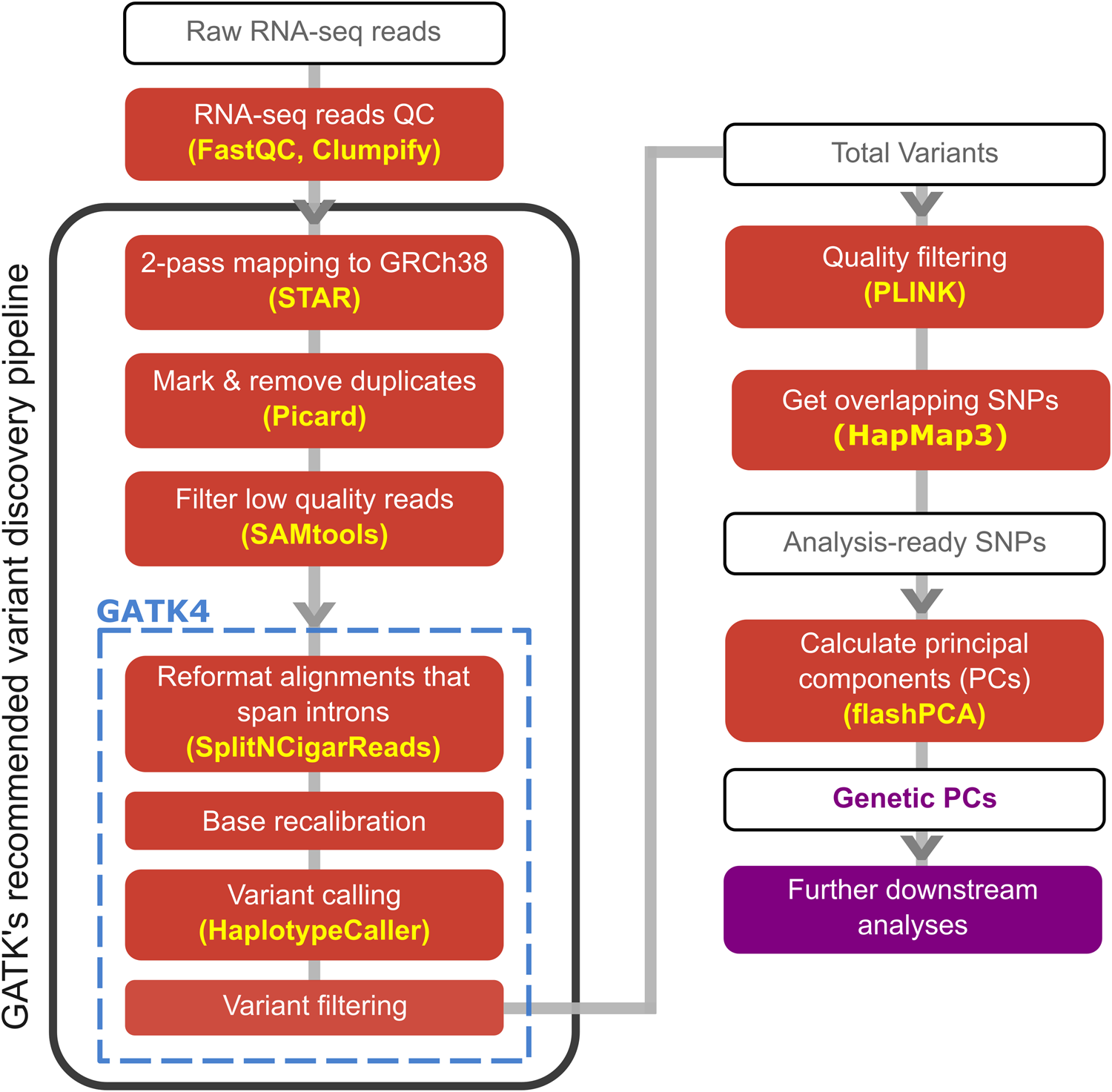
The analysis pipeline used in this study, mainly following GATK’s recommended variant discovery pipeline to call genetic variants from RNAseq samples
Further filtering based on missingness, MAF, and LD was then done before generating principal components (PCs) representing the population structure.
Availability – https://github.com/fachrulm/RGStraP
Fachrul M, Karkey A, Shakya M, Judd LM, Harshegyi T, Sim KS, Tonks S, Dongol S, Shrestha R, Salim A; STRATAA study group; Baker S, Pollard AJ, Khor CC, Dolecek C, Basnyat B, Dunstan SJ, Holt KE, Inouye M. (2023) Direct inference and control of genetic population structure from RNA sequencing data. Commun Biol 6(1):804. [article]