Single-cell multiomics can provide comprehensive insights into gene regulatory networks, cellular diversity, and temporal dynamics. While tools for co-profiling the single-cell genome, transcriptome, and epigenome are available, accessing the proteome in parallel is more challenging. To overcome this limitation, researchers at the Pacific Northwest National Laboratory have developed nanoSPLITS (nanodroplet SPlitting for Linked-multimodal Investigations of Trace Samples), a platform that enables unbiased measurement of the transcriptome and proteome from same single cells using RNA sequencing and mass spectrometry-based proteomics, respectively. The researchers demonstrated the nanoSPLITS can robustly profile > 5000 genes and > 2000 proteins per single cell, and identify cell-type-specific markers from both modalities.
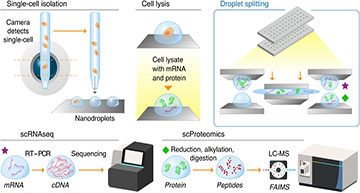
Overview of the nanoSPLITS-based single-cell multiomics platform
Schematic illustration showing the workflow including cell sorting, lysis, droplet merging/mixing, and droplet separation for downstream scRNAseq and scProteomics measurement.
Fulcher JM, Markillie LM, Mitchell HD, Williams SM, Engbrecht KM, Moore RJ, Cantlon-Bruce J, Bagnoli JW, Seth A, Paša-Tolić L, Zhu Y. (2023) Parallel measurement of transcriptomes and proteomes from same single cells using nanodroplet splitting. bioRXiv [online preprint]. [article]