The random noises, sampling biases, and batch effects often confound true biological variations in single-cell RNA-sequencing (scRNA-seq) data. Adjusting such biases is key to the robust discoveries in downstream analyses, such as cell clustering, gene selection and data integration. Researchers at Xiamen University have developed a model-based downsampling algorithm based on minimal unbiased representative points (MURPXMBD). MURPXMBD is designed to retrieve a set of representative points by reducing gene-wise random independent errors, while retaining the covariance structure of biological origin hence provide an unbiased representation of the cell population. Subsequent validation using benchmark datasets shows that MURPXMBD can improve the quality and accuracy of clustering algorithms, and thus facilitate the discovery of new cell types. Besides, MURPXMBD also improves the performance of dataset integration algorithms. In summary, MURPXMBD serves as a useful noise-reduction method for single-cell sequencing analysis in biomedical studies.
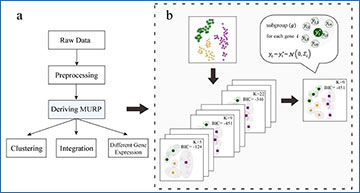
A schematic representation of MURP
(a) MURP is applied to preprocessed scRNA-seq data before downstream analyses of different purposes; (b) MURP is defined as an optimal set of representative points of the original cell population by iterative minimization of pseudo-BIC.
Availability – R implementations of MURP are available on GitHub (https://github.com/renjun0324/MURP) for academic use.
Ren J, Zhang Q, Zhou Y, Hu Y, Lyu X, Fang H, Yang J, Yu R, Shi X, Li Q. (2022) A downsampling method enables robust clustering and integration of single-cell transcriptome data. J Biomed Inform [Epub ahead of print]. [article]