Understanding how cells function and how diseases develop at a molecular level is a major goal in biology and medicine. Recently, researchers at the Huazhong Agricultural University have developed a new technology called MiP-seq, offering exciting advancements in this field. MiP-seq, short for multi-omics in situ pairwise sequencing, allows scientists to study various biological molecules like DNA, RNA, proteins, and other biomolecules all at once, and at a very detailed, subcellular level.
MiP-seq is a powerful technique that combines multiple types of molecular analysis into one comprehensive method. Traditional methods often focus on one type of molecule at a time, but MiP-seq can detect and analyze several types simultaneously. This gives researchers a more complete picture of what’s happening inside cells. The “in situ” part of MiP-seq means that the analysis is done directly in the tissue where the cells are located, rather than requiring the cells to be removed and processed separately. This helps preserve the natural context and spatial relationships within the tissue, which is crucial for understanding how cells interact with each other.
MiP-seq involves several key steps. First, tissue samples are prepared in a way that preserves their structure. The method then uses advanced techniques to label and detect DNA, RNA, proteins, and other biomolecules within these samples. MiP-seq decodes the molecular information through sequencing and imaging, generating detailed maps of where different molecules are located within the tissue.
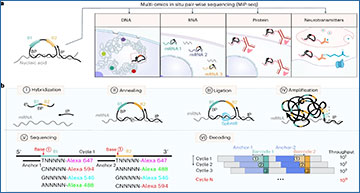
Application of MiP-seq for the spatial profiling of biomolecules
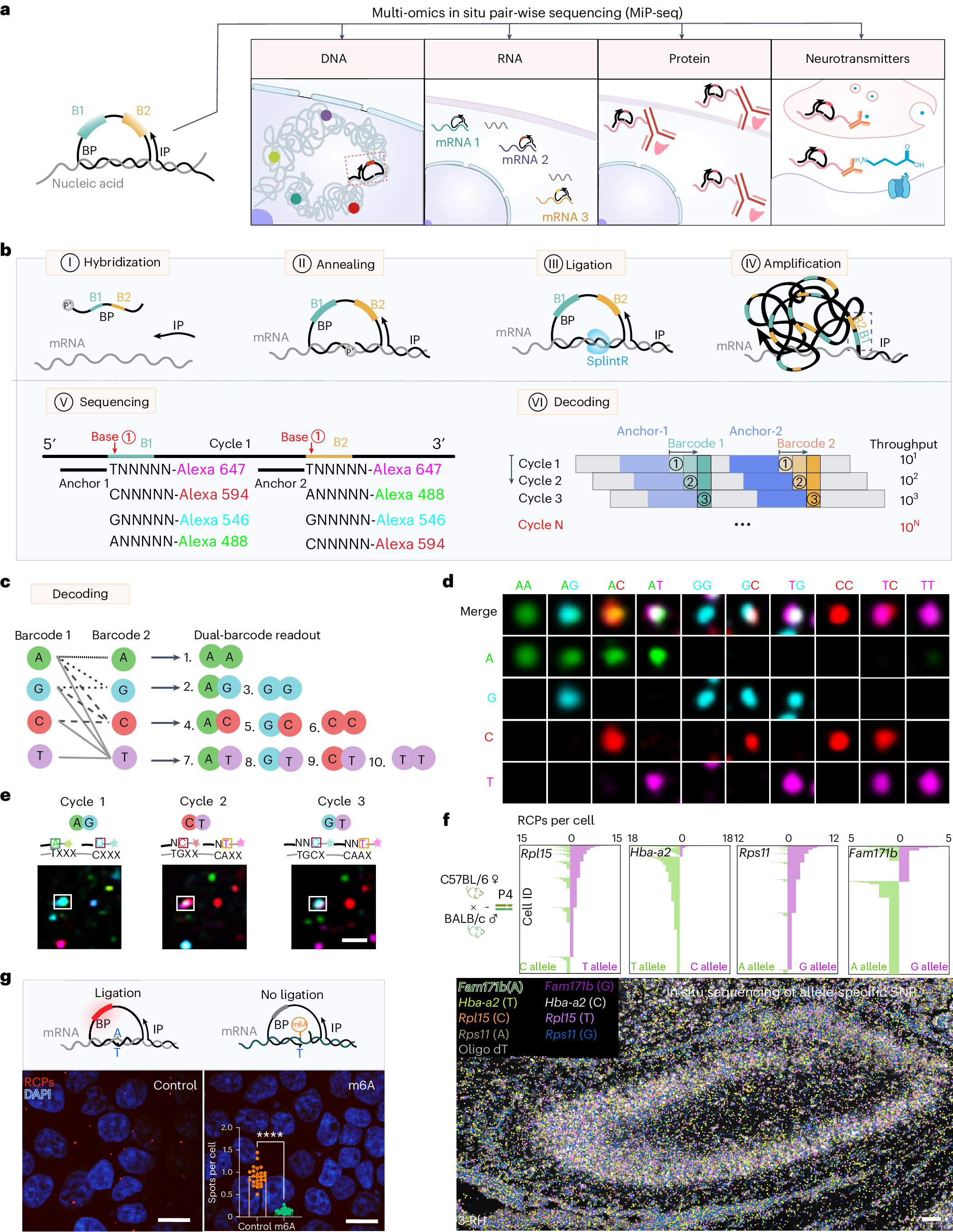
a, Diagram for the application of MiP-seq in spatial-omics profiling. b, Flow diagram of MiP-seq for RNA in situ detection. I. The dual barcode primer (BP) and initiator primer (IP) are hybridized to the target RNA. II. BP and IP are annealed to the target mRNA to form a padlock structure. III. BP is circularized by RNA-dependent DNA ligase SplintR to form the RCA template. IV. RCA of circled BP forms rolling-circle products. V. The dual-anchor primers are hybridized next to the dual-barcode sequence. Subsequently, the barcode sequences are interrogated by fluorescence-labelled query probes through sequencing by ligation. VI. Ten dual-barcode readouts are decoded in each round of sequencing, and 10N codes can be interrogated after N rounds. c, Illustration of ten dual barcodes interrogation according to merged fluorescence signals from paired bases for each round of sequencing.The thin dotted line indicates the paired bases AA. The thick dotted line indicates the paired bases AG and GG. The dashed line indicates the paired bases AC, GC and CC. d, Merged fluorescence signals for each of ten dual barcodes. e, Example of dual-barcode sequence interrogation. The boxed dots show the corresponding barcode reading for each of the three rounds. f, Bottom: in situ sequencing for allele-specific gene expression of Fam171b, Hba-a2, Rpl15 and Rps11 in hippocampus of offspring from C57BL/6 female mice crossed with BALB/c male mice. Top: expression level (RCPs per cell) of Fam171b, Rps11, Hba-a2 and Rpl15 alleles for each cell (n = 6,084 cells) in hippocampus. RH, right hippocampus. Cell morphology stained with oligo-dT-Cy3 (grey) was overlaid with corresponding gene-expression profiles. The detection of allele-specific gene expression was replicated at least three times. g, The m6A-methylated RNA and the unmethylated RNA were transfected into HEK-293T cells, which can be differentiated by MiP-seq. Statistical analysis was performed using Student’s t-test (two-tailed) for unmethylated and methylated RNA from 25 imaging fields (data are presented as mean ± s.d., ****P < 0.0001; the raw data can be found in Supplementary Table 3). The RCPs (Cy3 red fluorescence) and DAPI (blue fluorescence) are shown. The detection of m6A-methylated RNA and the unmethylated RNA was replicated at least three times. Scale bars, 2 µm in e, 50 µm in f, 15 µm in g.
MiP-seq offers several advantages. It can process a large amount of molecular data at once, making it very efficient. It also reduces the overall costs of sequencing and imaging compared to other similar techniques. Additionally, it maintains high efficacy in detecting specific genetic mutations, differences in gene expression, and modifications in RNA.
One of the exciting applications of MiP-seq is in the study of brain tissues. Researchers have used it to create a “spatial multi-omics atlas” of mouse brains. This means they can map out where different genes are active and how this correlates with neuronal activity and other cellular processes.
MiP-seq can be combined with other imaging techniques, such as calcium imaging, which allows scientists to see how active neurons are by tracking calcium ions, which are crucial for nerve cell signaling. It can also be combined with Raman imaging, which provides information about the biochemical composition of cells, giving further insights into cellular functions and interactions.
One challenge in in situ sequencing is dealing with “optically crowded” signals, where too many molecules are packed into a small space, making it hard to distinguish them. MiP-seq addresses this with a “sequential dilution strategy,” which helps separate these signals for clearer analysis.
The development of MiP-seq represents a significant step forward in our ability to analyze tissues at a molecular level. Its ability to provide a multidimensional view of cellular and molecular landscapes could lead to new discoveries in both basic biology and disease mechanisms. As this technology continues to evolve, it holds promise for advancing research in various fields, including neuroscience, cancer research, and regenerative medicine.
MiP-seq is a groundbreaking tool in spatial multi-omics that allows for comprehensive, cost-effective, and accurate analysis of multiple types of biomolecules within tissues. Its integration with other advanced imaging techniques opens up new possibilities for understanding the complex interactions that drive cellular functions and disease processes.
Wu X, Xu W, Deng L, Li Y, Wang Z, Sun L, Gao A, Wang H, Yang X, Wu C, Zou Y, Yan K, Liu Z, Zhang L, Du G, Yang L, Lin D, Yue J, Wang P, Han Y, Fu Z, Dai J, Cao G. (2024) Spatial multi-omics at subcellular resolution via high-throughput in situ pairwise sequencing. Nat Biomed Eng [Epub ahead of print]. [article]