We often hold it to be self-evident that antibodies are key to an effective immune response against a pathogen—the more (or more potent) antibodies your body makes, the better it will fight off an infection. What if I told you that—in certain situations—having antibodies against a pathogen can actually make an infection worse? This phenomenon is real, and it goes by the official name of antibody-dependent enhancement (ADE). One particular disease in which ADE is particularly problematic is infection with dengue virus, a flavivirus of major global health concern. While we still don’t fully understand the mechanisms behind ADE, we do know that it appears to be mediated by non-neutralizing antibodies—those that bind viral particles but paradoxically help the virus enter our cells, making the infection worse than if those antibodies hadn’t been there in the first place.
Still don’t believe me? Take it from Dr. Jay Lubow, a former post-doctoral fellow in Dr. Leslie Goo’s Lab in Fred Hutch’s Vaccine and Infectious Disease Division.
“There are four main types of dengue virus,” begins Dr. Lubow. “If you get infected with one type, chances are your immune system will produce two main categories of antibodies: those that can potently ‘neutralize’ the specific type of dengue you were exposed to, and those thatcan recognize but only poorly neutralize all four virus types. Sometimes, your body can also produce antibodies which can potently neutralize all four virus types, which we refer to as broadly-neutralizing antibodies (bnAbs)—these are more likely to be produced after 2 or more sequential exposures to different types. Now, if you later get infected with a different dengue virus type, any remaining cross-reactive but poorly neutralizing antibodies can mediate ADE and worsen your illness.”
As you might imagine, this phenomenon is a major challenge for designing dengue vaccines, which must minimize the risk of ADE if they are to safely and effectively curb infection. Accordingly, a major focus of dengue virus research is on the aforementioned bnAbs. What differentiates them from other antibodies? What determines whether a vaccine preferentially elicits bnAbs, and how can we promote this process? Of course, to better understand bnAbs against a specific pathogen, we must first identify them; this is where the Goo Lab’s recent publication comes in.
“Before this study, there were a few known bnAbs against dengue virus and the related Zika virus, and they were mostly of a similar type.” notes Dr. Lubow.
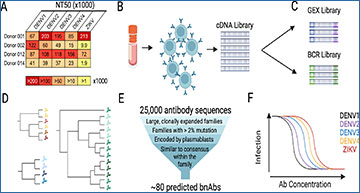
Part of the reason so few bnAbs were known is due to the generally laborious task of isolating them. Traditionally, scientists had to find patients who had been infected—ideally, multiple times—with dengue virus, collect their blood, isolate individual B cells (the antibody-producing immune cells), expand them in culture and then purify the antibodies they secrete. Each B cell produces a unique antibody, whose sequence is encoded in its DNA. Thus, an alternative approach to identify antibodies bypasses the B cell culturing and instead directly uses polymerase chain reaction (PCR) to amplify the antibody-encoding regions of sorted B cells. However, these methods are labor intensive and require a priori knowledge of the antibodies or antibody types one is looking for.
“Our recent publication describes an effort to find new bnAbs against dengue virus using a novel approach that we have been developing for a few years,” says Lubow.
While the initial steps of the team’s antibody hunt are similar to previous approaches—they still needed to isolate B cells from patients with multiple dengue virus infections—they opted to hunt for bnAbs using single cell RNA sequencing. The ability to obtain antibody RNA sequences and map these sequences back to the single B cells from which they came was not only higher-throughput than previous approaches; it also allowed them to detect comparatively rare antibody types in an unbiased manner. Collaborating with members of the Matsen Lab in the Public Health Sciences Division, the team employed computational algorithms to group antibody sequences into families based on sequence identity. They then used this grouping to select representative antibody types and experimentally screen them for their ability to broadly neutralize a collection of dengue and Zika viruses. All in all, their efforts were handsomely rewarded: using their method, Lubow and colleagues discovered 23 new bnAbs against dengue (and Zika, in some cases), some of which rivaled or even outperformed the most potent bnAbs previously discovered!
To fully appreciate the significance of the Goo Lab’s innovation, it’s important to note that until this study, all of the leading dengue virus bnAbs were of a single antibody class known as immunoglobulin G (IgG). This is somewhat peculiar when you consider that humans have significant amounts of other antibody classes, but there’s a rather simple explanation: the methods used to discover these antibodies (mentioned above) often specifically sort or enrich for IgG. While most of the new bnAbs that Lubow and team discovered using their unbiased approach were also IgG, they were surprised to find that their highest-performing bnAb was of the IgA class—the first such bnAb against dengue virus ever identified. Not only that, but when the team tested this IgA bnAb, they made another shocking discovery: even at sub-neutralizing concentrations, this bnAb did not exhibit ADE in vitro, and was even able to suppress ADE when mixed with IgG antibodies that normally enhance infection!
A plot with percent infection on the y-axis and antibody concentration on the x-axis illustrates suppression of antibody-dependent enhancement (ADE) by IgA antibodies against dengue virus. In a sample containing only IgG antibodies (top, dark green line), non-neutralizing concentrations of antibody lead to enhancement of infection via ADE, giving a characteristic bell-shaped curve. This effect disappears in mixtures containing sequentially larger proportions of IgA antibody (lighter green lines) and is absent in conditions with only IgA antibody (bottom line). Image taken from publication.
“Overall, we were quite surprised with the amount of bnAbs that we were able to characterize using our unbiased method,” notes Lubow,“ and our discovery of an IgA against dengue that combats antibody-dependent enhancement really places IgAs, which have largely been overlooked in favor of IgGs, as potentially high-interest targets for safe and effective dengue vaccine design.”
In terms of future directions, this study will keep researchers busy for years to come. Are there other IgA bnAbs against dengue and Zika viruses out there to discover? Do IgA antibodies against dengue inhibit ADE in vivo, and, if so, by what mechanism? Can the rich antibody transcriptome datasets generated using this approach be co-opted to study antibody evolution or inform computational antibody design? To those of us on the hunt for our own discoveries (scientific or otherwise), this story also reminds us that we usually find the types of things we set out to look for. Sometimes, big discoveries can be made not by looking harder, but by looking for things that others have overlooked!
Source – Fred Hutch Cancer Center
Lubow J, Levoir LM, Ralph DK, Belmont L, Contreras M, Cartwright-Acar CH, et al. (2023) Single B cell transcriptomics identifies multiple isotypes of broadly neutralizing antibodies against flaviviruses. PLoS Pathog 19(10): e1011722. [article]