Researchers have demonstrated that, despite the intrinsically low raw accuracy of long-read sequencing technologies, multiplex sequencing in these technologies can be improved to a read recovery rate and crosstalk rate similar to current high-accuracy short-read sequencing technologies using non-random error-correcting barcodes synthesized in massive parallel on a microarray.
The results of the study have been published in Scientific Reports.
Next generation sequencing technologies are revolutionizing genomics research. In particular, long-read sequencing holds the promise of fully characterizing more challenging genomes at high resolution. However, this emerging technology suffers from reduced raw accuracy when compared to current short-read sequencing technologies. Random DNA barcodes have already been successfully used in high-accuracy short-read sequencing as unique molecular identifiers to filter out duplicate reads and PCR errors. Error-correcting non-random DNA barcodes represent a possible solution to improve recovery in spite of the low raw accuracy of long-read sequencing. These barcodes of known sequence contain error-correcting codes inspired by watermark error-correcting codes that were originally developed to deal with the issue of reliable transmission of information in the presence of noise in digital communications. This systematic approach makes it possible to design an arbitrary number of distinct barcodes.
However, synthesis of such a large number of known sequences using traditional column-based synthesis methods is cost prohibitive. Researchers at CONICET, the National University of Rosario (UNR), CCTyE Acuario del Río Paraná and The Hebrew University of Jerusalem have investigated the feasibility of using LC Sciences’ OligoMix® oligonucleotide pools to provide the larger number of DNA barcode sequences required and improve the accuracy of the long-read sequencing. OligoMix® is produced via a patented microfluidic massively parallel synthesis technology developed at LC Sciences.
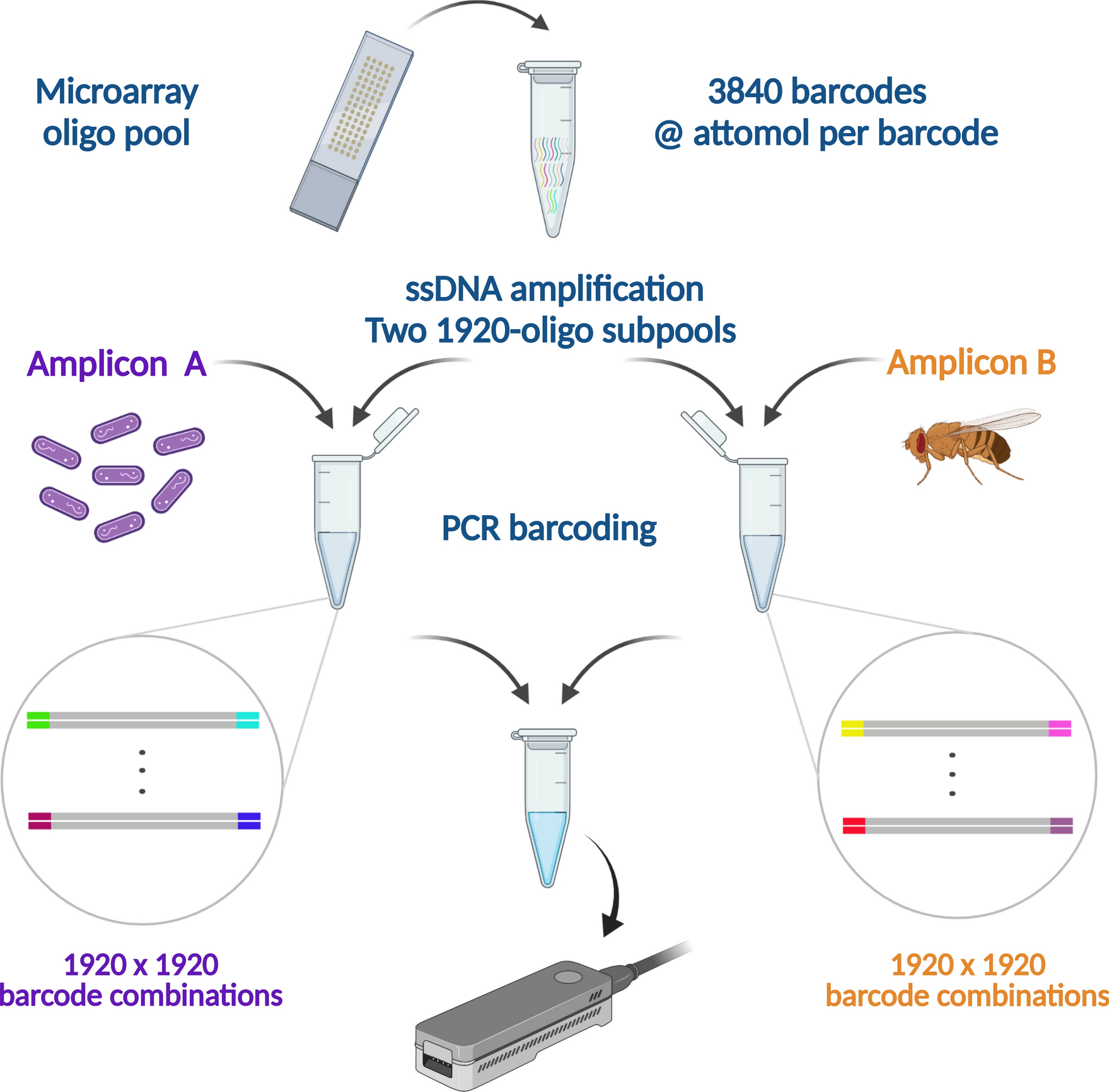
A research team led by Dr. Joaquín Ezpeleta and Dr. Elizabeth Tapia designed 3840 distinct 76-bp oligos that each contained a 36-bp variable barcode sequence flanked by two sets of fixed, 20-bp primer annealing regions used to amplify two subpools A and B of 1920 oligos. These subpools were used as primers in two separate barcoding PCR reactions and then the barcoded products were subjected to standard library preparation and long-read sequencing.
A barnyard experiment (two species) is used to assess the feasibility and performance of NS-watermark barcoding for massively parallel sequencing
A subset of 3840 NS-watermark barcodes, from a set of 4096, is synthesized in a microarray and cleaved to form an oligo pool, which is further divided into two subpools of 1920 barcodes each, followed by preferential amplification of one strand. The resulting ssDNA-enriched subpools are used to tag B. Pertussis (sample A) and D. mojavensis (sample B) amplicons. Barcoded amplicon subpools A and B are pooled in a molar ratio of 1:2 and sequenced together on the MinION sequencing platform.
The researchers reported a read recovery rate of 86.4% at a crosstalk rate of 0.17% (≈ 1 misassignment for every 584 demultiplexed reads). Despite the higher plexity and the combined error rate of microarray-based synthesis, multiple rounds of PCR amplification using low-fidelity Taq polymerase and native single-pass Oxford Nanopore Technologies (ONT) sequencing, the resulting level of crosstalk is comparable to that exhibited by 96-plex single-end barcoding schemes on a short-read platform.
Additionally, the researchers reported full representation of the microarray-synthesized oligonucleotides: all 3840 barcodes in subpools A and B were detected in at least 10 of the reads, while none of their 256 negative control barcodes were detected in more than 5 reads. Similarly, rich diversity was observed in terms of barcode combinations.
According to the research team, the number of distinct barcodes synthesized and demultiplexed in their study is one order of magnitude higher than the largest commercially-available barcoding kits for long-read sequencing (3840 vs. PacBio’s 384), and the diversity afforded by this many distinct tags with asymmetric barcoding would be close to that offered by 12-nt unique molecular identifiers. The researchers suggest that even higher numbers are within reach, as the NS-watermark barcode design framework is arbitrarily scalable (limited only by barcode length) and microarray-based synthesis of tens of thousands of oligonucleotides per chip or more is already commercially available.
LC Sciences routinely synthesizes oligos at double the length used here (up to 150-bp) in batches containing up to 30,000 unique known sequences.
Several members of the research team founded a biotech startup, ArgenTAG Corp., to develop barcoding and single-cell sequencing kits for long-read sequencers based on this technology.
Dr. Ezpeleta, now Director of Engineering at ArgenTAG, commented: “We had been working on the design of large families of error tolerant barcode sequences for a while, and sequencing throughput has been steadily increasing over the years. OligoMix was the missing piece: a reliable and cost-effective way of synthesizing thousands of oligos. The combined ability to design, ‘write’ and ‘read’ barcode oligos at scale opens a whole new range of possibilities”.
Chris Hebel, Vice President of Business Development at LC Sciences commented, “It’s really exciting for us when our customers develop innovative new applications for OligoMix. It’s a complex product, with many thousands of sequences pooled together in such a small scale. Generally, it requires very creative thinking to envision its full potential in new applications. The CONICET research team’s inventive design and execution of their project exemplify this.”
Source – LC Sciences
Ezpeleta J, Garcia Labari I, Villanova GV, Bulacio P, Lavista-Llanos S, Posner V, Krsticevic F, Arranz S, Tapia E. (2022) Robust and scalable barcoding for massively parallel long-read sequencing. Sci Rep 12(1):7619. [article].