Full-length RNA-sequencing methods using long-read technologies can capture complete transcript isoforms, but their throughput is limited. Researchers at the Broad Institute of MIT and Harvard have developed multiplexed arrays isoform sequencing (MAS-ISO-seq), a technique for programmably concatenating complementary DNAs (cDNAs) into molecules optimal for long-read sequencing, increasing the throughput >15-fold to nearly 40 million cDNA reads per run on the Sequel IIe sequencer. When applied to single-cell RNA sequencing of tumor-infiltrating T cells, MAS-ISO-seq demonstrated a 12- to 32-fold increase in the discovery of differentially spliced genes.
MAS-ISO-seq workflow and experimental validation using synthetic RNA isoforms
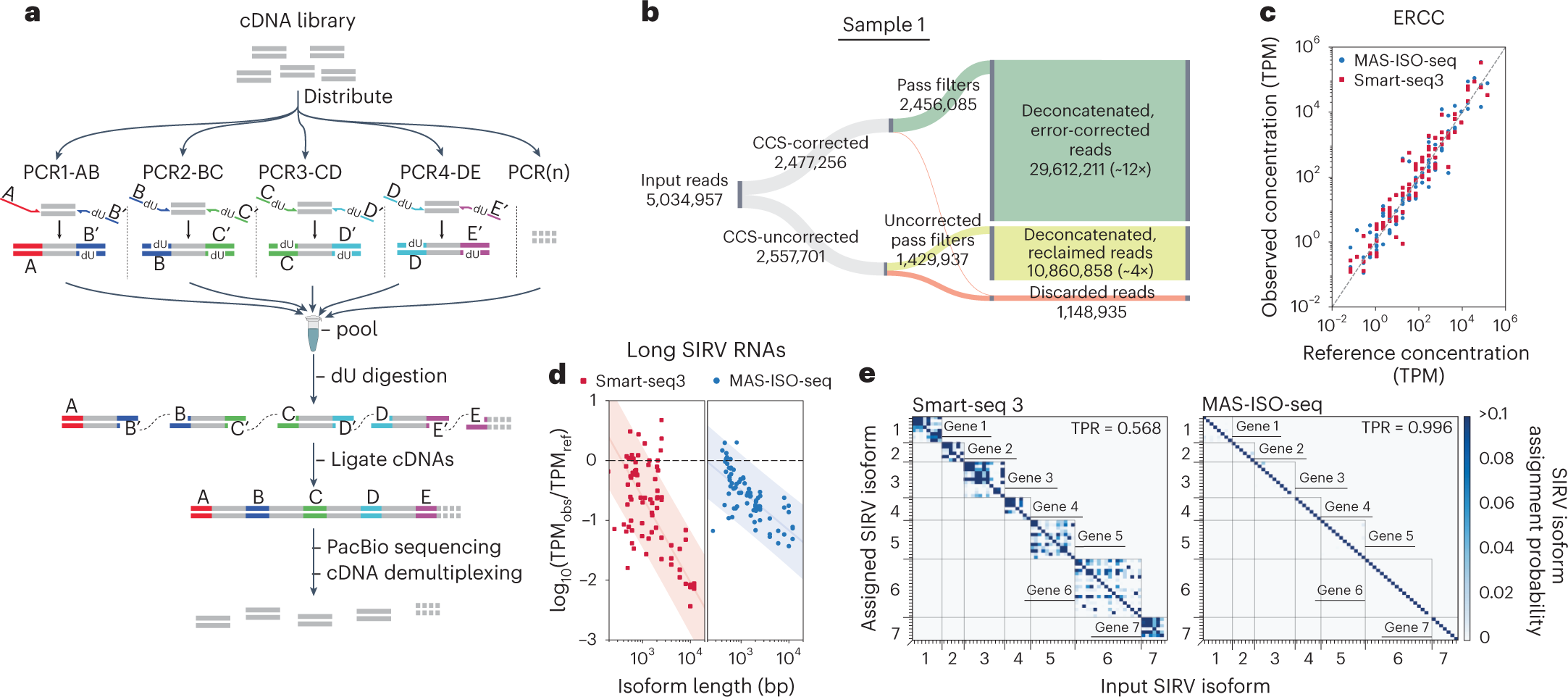
a, Schematic representation of the MAS-ISO-seq intramolecular cDNA multiplexing workflow. b, Sankey diagram reporting MAS-ISO-seq run yield of sample 1 at various stages of processing. c, Observed ERCC concentrations as measured in MAS-ISO-seq and Smart-seq3 experiments versus reference concentrations (R2 > 0.95 for both). d, Log ratio of observed to reference concentrations of short and long SIRV isoforms in SIRV-Set 4 versus transcript length for Smart-seq3 and MAS-ISO-seq. e, Isoform identification confusion matrix for SIRV isoforms as measured by Smart-seq3 reconstructions and MAS-ISO-seq observations.
Al’Khafaji AM, Smith JT, Garimella KV, Babadi M, Popic V, Sade-Feldman M, Gatzen M, Sarkizova S, Schwartz MA, Blaum EM, Day A, Costello M, Bowers T, Gabriel S, Banks E, Philippakis AA, Boland GM, Blainey PC, Hacohen N. (2023) High-throughput RNA isoform sequencing using programmed cDNA concatenation. Nat Biotechnol [Epub ahead of print]. [abstract]