RNA molecules are the messengers that convey crucial information within cells, orchestrating various biological processes. However, these RNA molecules are not static entities; they undergo modifications that add an additional layer of complexity to the transcriptome.
One such modification, known as m6A methylation, plays a significant role in regulating gene expression and ultimately influencing cellular functions. Detecting these RNA modifications is essential for understanding their impact on cellular processes and their potential implications for health and disease.
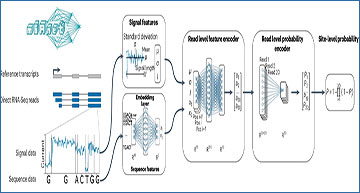
In a groundbreaking study, researchers at the National University of Singapore have developed a cutting-edge method called m6Anet, which leverages the power of neural networks and machine learning to unravel the mysteries of RNA modifications. Specifically designed to analyze data from nanopore direct RNA sequencing, m6Anet can capture the raw current signal for each RNA molecule, enabling the detection of m6A modifications with unprecedented accuracy.
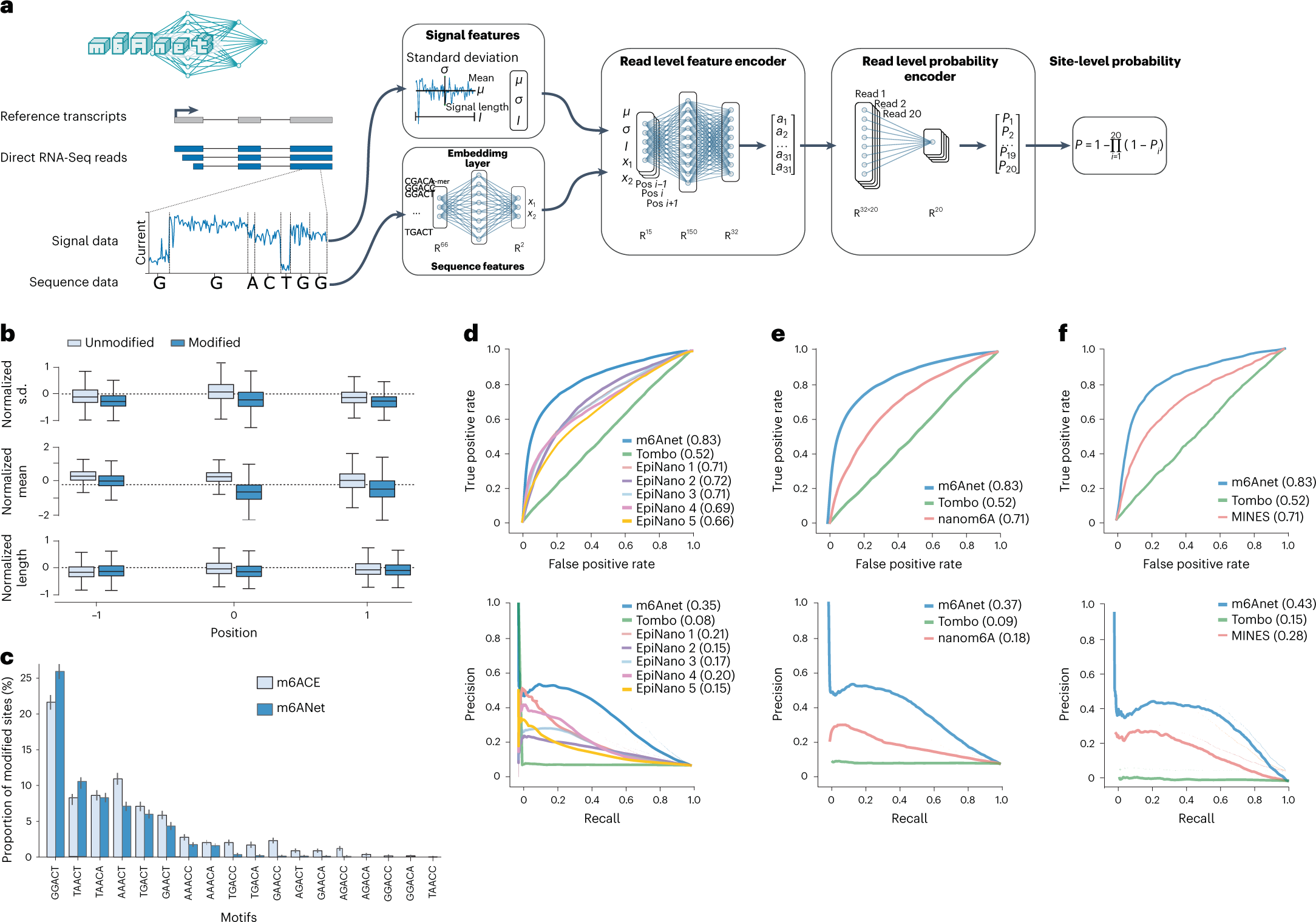
Schematic of m6Anet and evaluation on detection of m6A in human cell lines
a, m6Anet model schematics. b, Box plot showing the difference in average features distribution between different m6Anet prediction with n = 1769 predicted modified sites and n = 5031 predicted unmodified sites for the GGACT fivemer motif. The horizontal lines on the boxes show minima, 25th percentile, median, 75th percentile, and maxima. Points that do not fall within 1.5× of the interquartile range are considered outliers and are not shown on the plot. c, Comparison of the proportion of modified sites predicted as modified by m6Anet and by m6ACE on the DRACH fivemer motifs. The bar plot center represents the proportion of modified sites for each fivemer motif while the error bar represents the estimated 95% confidence interval around the center values with a total of n = 5,579 for m6ACE modified positions, n = 4,784 for m6Anet-predicted modified positions and n = 121,853 for m6ACE unmodified positions and n = 122,648 for m6Anet-predicted unmodified positions. d, ROC curve (top) and PR curve (bottom) of m6Anet against all five EpiNano models and Tombo. e, ROC curve (top) and PR curve (bottom) of m6Anet against nanom6A and Tombo. f, ROC curve (top) and PR curve (bottom) of m6Anet against MINES and Tombo.
One of the key challenges in analyzing RNA modifications is the lack of complete data at the single RNA molecule level. Experimental approaches often provide only site-level training data, leaving gaps in our understanding of the modification status for individual RNA molecules. m6Anet addresses this challenge by employing a sophisticated multiple instance learning framework, allowing it to handle missing read-level modification labels in site-level training data effectively.
m6Anet outperforms existing computational methods and demonstrates similar accuracy to experimental approaches in identifying m6A modifications. Furthermore, it exhibits remarkable generalization capabilities, accurately detecting RNA modifications across different cell lines and species without the need for retraining model parameters.
One of the most exciting aspects of m6Anet is its ability to capture the underlying read-level stoichiometry of RNA modifications. This means that it can not only identify the presence of m6A modifications but also provide insights into the relative abundance of these modifications within RNA molecules. This information is invaluable for understanding differences in modification rates and their potential functional consequences.
Overall, m6Anet represents a significant advancement in the field of transcriptome analysis. By offering a comprehensive tool for the identification and quantification of m6A modifications from a single run of direct RNA sequencing, m6Anet opens up new possibilities for studying RNA biology and its role in health and disease.
Availability -The source code for m6Anet is available at https://github.com/GoekeLab/m6anet.
Hendra C, Pratanwanich PN, Wan YK, Goh WSS, Thiery A, Göke J. (2024) Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat Methods 19(12):1590-1598. [article]