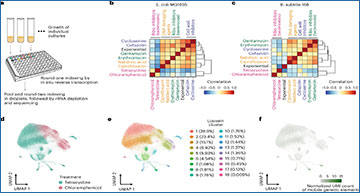
Three Princeton laboratories teamed up to develop a method for studying gene expression patterns of individual bacteria in unprecedented detail.
Bacteria are nearly ubiquitous and have tremendous impacts on human and ecological health. And yet, they remain largely mysterious to us. Princeton MOL faculty Zemer Gitai, Britt Adamson and Ned Wingreen launched a joint effort to develop new tools to help us better understand bacteria.
Bacteria are incredibly numerous and astoundingly diverse. The human gut contains as many bacterial cells as there are human cells in the entire body, spread across an estimated 500-1000 bacterial species. Bacteria also colonize surfaces and organisms all around us.
Individual bacteria of the same species may appear superficially similar but still exhibit diverse responses to stress, depending on the expression pattern of their genes—that is, which genes are actively being used to make end products such as proteins that affect cell behavior. Historically, this type of variability among cells was hard to capture because the tools used to study bacteria were only suited to making generalizations about population-wide trends in gene expression. This obscured differences in gene expression that could explain important behaviors such as development of antibiotic resistance.
Recently, scientists have begun leveraging next-generation sequencing and associated techniques, such as single-cell RNA sequencing, that researchers can use to study gene expression in hundreds of thousands of individual cells simultaneously. However, many challenges remain. For example, some of these new approaches are limited to studying only a narrow set of genes that we already know about, while others are hampered by technical issues that impair their sensitivity. The Princeton team, spearheaded by graduate student Bruce Wang, set out to overcome these limitations.
In order to observe which genes are being expressed in a cell, scientists need to look for a type of molecule called messenger RNA (mRNA for short), which appears when a protein-coding gene is being expressed. The problem arises because up to 97% of a bacterium’s RNA molecules are a different type of RNA, called ribosomal RNA (rRNA). Most approaches work by first tagging a random selection of all RNA molecules from the cell and then using deep sequencing to discover the tagged molecules’ identities. The more abundant rRNA gets sequenced more often than mRNA, resulting in poor signal-to-noise ratios. Wang and his colleagues developed a way to remove ribosomal RNA sequences after the tagging step but before the sequencing step, which both enriches mRNA in the sample and reduces the expense of sequencing. Wang also adopted a barcoding-based method to allow this approach to be applied to hundreds of thousands of cells at once.
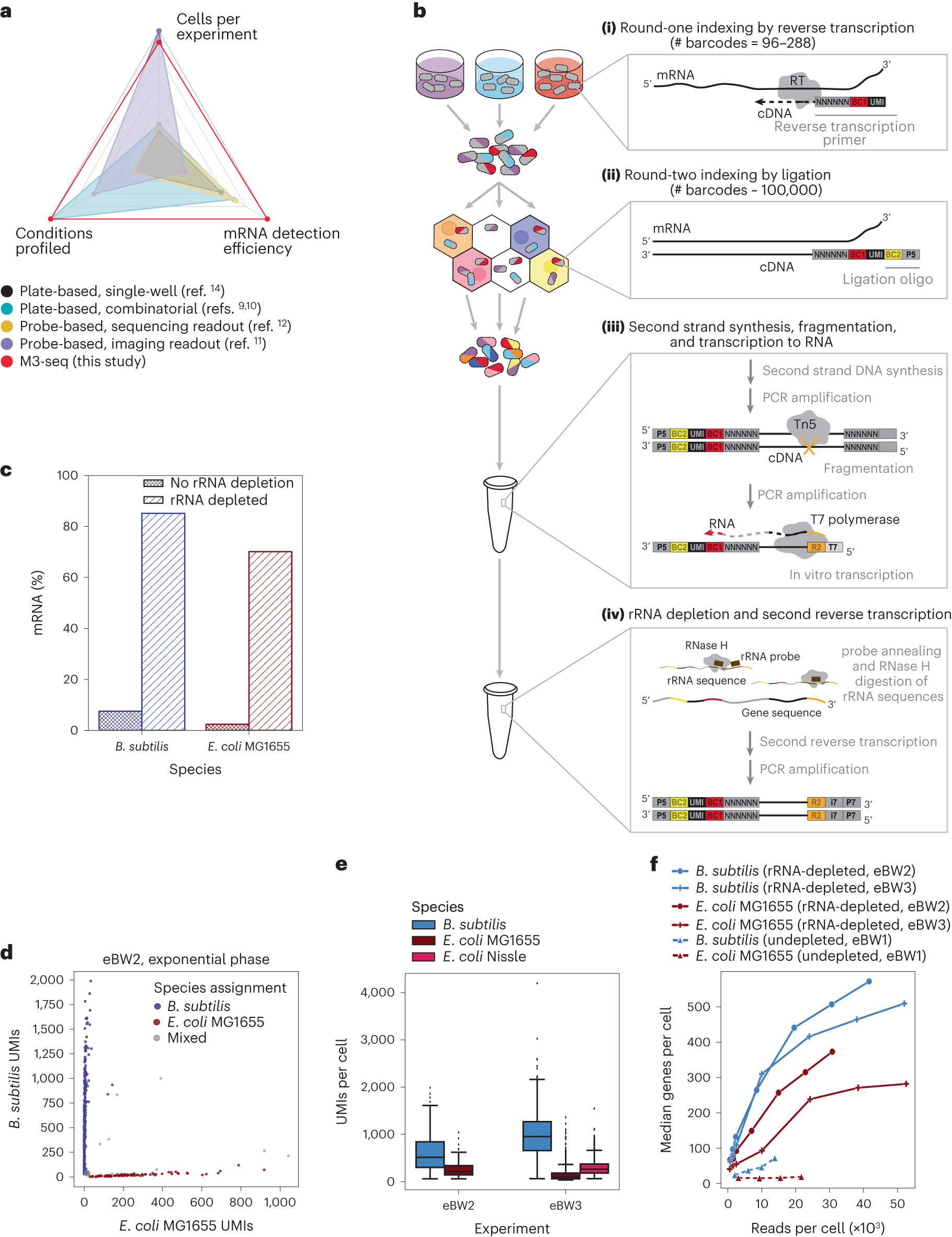
Development of M3-seq platform for single-cell RNA-sequencing with post hoc rRNA depletion
a, scRNA-seq methods previously established for bacteria with reported number of cells (ranging from 100 cells per experiment to 300,000 cells per experiment), conditions (ranging from 1 condition per experiment to 20 conditions per experiment) and mRNA genes per cell (ranging from 29 genes per cell to 371 genes per cell). Numbers in each category were selected by taking maximum reported values. Numbers also found in Extended Data Table 1. b, Schematic of M3-seq experimental workflow. Indexing: (i) RNA molecules are reverse transcribed in situ with indexed primers such that cells in each reaction (that is, separate plate wells) are marked with distinct sequences. Primers allow for random priming. (ii) Cells are then collected, mixed and distributed into droplets for a second round of indexing via ligation with barcoded oligos. Sequencing library preparation: Cells are collected again and lysed to release single-strand cDNAs. (iii) Second-strand synthesis is then performed in bulk reactions and resulting cDNA molecules are fragmented with Tn5 transposase, amplified via PCR to add a T7 promoter and converted to RNA using T7 RNA polymerase. (iv) To deplete ribosomal sequences, the amplified RNA library is hybridized to complementary DNA probes (Supplementary Table 3), and annealed sequences are cleaved by RNase H. Finally, remaining sequences are reverse transcribed back to DNA, sequencing adaptors are added and data are collected by sequencing. c, Percentages of mRNA sequences in B. subtilis and E. coli single-cell libraries prepared with and without rRNA depletion. Data from undepleted libraries come from eBW1 and data from depleted libraries come from eBW3. d, M3-seq analysis of a mixture of B. subtilis and E. coli wherein each point corresponds to a single ‘cell’ (that is, unique combination of plate and droplet barcodes). Species assignments were made as described in Methods. e, UMIs per cell (after species assignment) observed in exponential-phase cells across two experiments, eBW2 and eBW3 (515 ± 245 and 953 ± 310 median UMIs with absolute deviation for B. subtilis, respectively; 211 ± 85 and 100 ± 47 median UMIs with absolute deviation for E. coli MG1655, respectively; 266 ± 100 UMIs with for E. coli Nissle in eBW3). N = 1,336, 533, 84, 1,944, 1,659 cells, respectively. Boxplot limits are as defined in Methods. f, Median genes detected per B. subtilis or E. coli cell as a function of the number of total reads per cell across three experiments: eBW1, eBW2 and eBW3.
Using this method, the Princeton group set out to investigate fundamental questions about bacterial biology. For example, they examined how E. coli bacteria respond to eight different types of antibiotics. As expected, different types of antibiotics evoked different responses, depending on the mode of action of the antibiotic. However, the responses of individual cells were remarkably varied, with different subpopulations expressing different subsets of genes. Further study of the genes involved could help scientists develop more effective antibiotics or antibiotic combinations targeting these genes.
The team also probed how bacteria respond to infections by a virus called bacteriophage λ. Surprisingly, they found that only a third of the bacteria actually become infected by λ and begin producing its proteins, even when the phage outnumbers bacteria by 100:1. This highlights how methods that only look at responses of a population as a whole can obscure potentially important biological complexities.

Examination of bacteria through M3-seq shows that only a subset of cells, represented by Cluster 3 in (top), actually become infected by bacteriophage λ and begin expressing viral genes. This finding was confirmed by using time-lapse microscopy, which showed that only about a third of cells are affected by virus (as shown by the dying cells, labeled red, in the time series at bottom). Figures courtesy of the authors
This exciting new technology, which the authors call M3-seq (short for “massively parallel, multiplexed, microbial sequencing”), is not limited to use on single species of bacteria. It can also be used to study multispecies communities of bacteria like those found in the human microbiome, opening the door to new realms of inquiry that the team is eager to explore.
Source – Princeton University
Wang B, Lin AE, Yuan J, Novak KE, Koch MD, Wingreen NS, Adamson B, Gitai Z. (2023) Single-cell massively-parallel multiplexed microbial sequencing (M3-seq) identifies rare bacterial populations and profiles phage infection. Nat Microbiol [Epub ahead of print]. [article]