Dr. Jorge Galeano Niño, a postdoctoral fellow in Dr. Susan Bullman’s Lab, confesses that he did not initially appreciate the importance of spatially probing microbe-driven transcriptional changes in cancers along the digestive system, an idea which resulted in his recent first-author Nature paper. An immunologist at heart, Galeano Niño says he was initially hooked on immunology after watching T cells through a microscope, moving in search of their target cell to kill, much like a hunter and its prey. Spatial transcriptomics might not instill the thrill of a live hunt, however Dr. Galeano Niño says that he quickly learned to appreciate the “power of spatial transcriptomics, from both a translational and basic science perspective.” This technique allowed him to look at how bacteria and tumor cells interact and whether microbes could influence cancer cell behavior. Microbiota are an important part of the tumor microenvironment in many cancers, especially those in the gut. However, since most studies focus on bulk tissue samples, potential localized roles that bacteria play in shaping the tumor microenvironment remain unknown. In collaboration with Dr. Christopher Johnston’s Lab, the Bullman research team investigated spatial variation in the ins and outs of the digestive system—oral and colorectal cancers—uncovering a surprising heterogeneity of microbial tumor distribution and their regional influence on epithelial and immune cell function that could enhance cancer progression.
The researchers first used spatial transcriptomic approaches to determine how tumor-associated microbes are distributed in these cancers. In both oral and colorectal patient tumor samples, Galeano Niño found that microbes reside in distinct niches, with the oncomicrobe Fusobacterium being one of the most dominant species in both cancer types, although it was found in greater abundance in colorectal tumors. Given the heterogenous distribution of intratumoral microbes within individual tumor tissues, the team then sought to understand what effect the bacteria had on the tumor microenvironment using a spatial profiling platform to analyze expression of proteins associated with anti-tumor immunity and cancer progression. The researchers found that bacteria reside in highly immunosuppressive microniches, with T cells being excluded from bacteria-colonized regions. Furthermore, they found a significant reduction in expression of the tumor suppressor, wild-type p53 at these sites, indicating that bacterial localization correlates with highly transformed cancer cells within the tumor microenvironment.
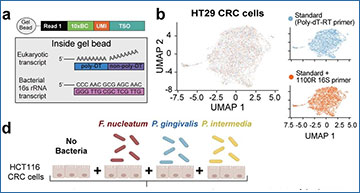
Next, the team asked how these host-bacterial interactions affect cellular function within the tumor microenvironment. To do so, they developed a method called INVADE-seq (invasion-adhesion-directed expression sequencing) which allowed them to apply a single cell RNA-seq approach that provided information about gene expression of the patient cancer cells in addition to identifying bacterial transcripts within these cells. Applying this technique to fresh oral cancer samples, they again found Fusobacterium among the dominant bacterial species present, primarily associated with epithelial and monocyte-derived macrophage cells in these patients. Epithelial cancer cells harboring bacteria had gene expression signatures enriched for signaling pathways involved in cancer progression, including epithelial-to-mesenchymal transition pathways and interferon response. Consistent with their spatial analysis, the researchers found the total bacterial load in cell clusters was negatively correlated with expression of wild-type TP53.
Are bacteria choosing to reside in places where they know they can thrive, or are they turning their region of residency into an immune-suppressive, cancer-driving niche? Galeano Niño admits this chicken or the egg relationship is difficult to determine and an ongoing question he is investigating. However, he was able to use some immunology tricks from his PhD work and developed functional in vitro assays to test if bacteria could cause changes in cancer cell behavior. Galeano Niño co-cultured colorectal epithelial cells with Fusobacterium nucleatum and embedded them in neutrophil-containing collagen matrices. Using live cell imaging and gene expression analyses, Galeano Niño uncovered that colorectal cancer cells harboring bacteria recruited neutrophils and resulted in an increase in cancer progression pathways including remodeling of extracellular matrix, cell adhesion and migration, while downregulating pathways involved in DNA damage repair and p53 signaling. Furthermore, he observed epithelial cells infected with bacteria detached from the spheroids and invaded the surrounding environment as single cells- a behavior reminiscent of metastatic behavior. These functional assays support the researchers’ findings in patient samples and suggest that bacteria are indeed capable of influencing cancer cell behavior to promote cancer progression.
Bacterial distribution in oral squamous cell carcinoma (OSCC) and colorectal cancer (CRC).
Collectively, this work uncovers that the distribution of microbiota within tumors is not random, but rather highly organized and potentially functionally-relevant. This study also highlights that there is much more heterogeneity within a tumor than originally appreciated. Galeano Niño points out that this could be particularly important when it comes to treatment options such as immunotherapies, which help ignite the tumor-fighting potential of immune cells. Such a therapy might help kill off the bacteria-free portion of the tumor but would likely have no effect on those immune-suppressive, highly transformed bacterial-colonized pockets. Galeano Niño emphasizes that their findings revealing the heterogeneity in these tumors “highlights the need to create more precise treatment regimens for each patient.”
Moving forward, Galeano Niño plans to look at different parts of the gut tract to understand how microbes might differentially affect cancers along the digestive system, in addition to investigating how intracellular vs. extracellular bacteria influence cancer cell function. Galeano Niño highlights the role his mentor, Dr. Bullman, played in helping shape this story and expand his view and appreciation for biology. He notes that “she challenged me in a good way and our different focuses—Susan’s on the translational aspect of this work and mine on finding functional ways to test the biology we were finding—synergized to expand the impact of this work.” Galeano Niño concludes with explaining that this work “taught me to appreciate the heterogeneity in tumors and recognize that this is a fundamental aspect of biology and also society. Heterogeneity is advantageous, since it enables biological systems to survive in rough environments; the T cells already know that—they know that they function better when they work together and collaborate. We can learn from biology here and learn how this collaboration benefits society too.”
Source – Fred Hutchinson Cancer Center
Availability – Custom code for data processing and analysis of 10x Visium spatial transcriptomics and scRNA-seq data is available at https://github.com/FredHutch/Galeano-Nino-Bullman-Intratumoral-Microbiota_2022.
Galeano Niño JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, Futran N, Houlton J, Sather C, Sicinska E, Taylor A, Minot SS, Johnston CD, Bullman S. (2023) Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611(7937):810-817. [article]