High-throughput total nucleic acid (TNA) purification methods based on solid-phase reversible immobilization (SPRI) beads produce TNA suitable for both genomic and transcriptomic applications. Even so, small RNA species, including miRNA, bind weakly to SPRI beads under standard TNA purification conditions, necessitating a separate workflow using column-based methods that are difficult to automate.
Researchers at the Michael Smith Genome Sciences Centre have developed an SPRI-based high-throughput TNA purification protocol that recovers DNA, RNA and small RNA, called GSC-modified RLT+ Aline bead-based protocol (GRAB-ALL), which incorporates modifications to enhance small RNA recovery. GRAB-ALL was benchmarked against existing nucleic acid purification workflows and GRAB-ALL efficiently purifies TNA, including small RNA, for next-generation sequencing applications in a plate-based format suitable for automated high-throughput sample preparation.
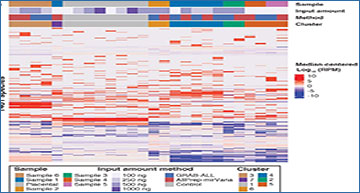
Workflow diagrams of TNA purification methods evaluated in this study
As depicted, AllPrep-mirVana workflow consists of two column-based protocols with tube-based steps that are difficult to automate (left). On the right, previously published TNA purification workflow based on Aline EvoPure was modified to enrich for small RNA (modified steps are denoted by *), giving rise to GRAB-ALL.
Xu J, Pandoh PK, Corbett RD, Smailus D, Bowlby R, Brooks D, McDonald H, Haile S, Chahal S, Bilobram S, Mungall KL, Mungall AJ, Coope R, Moore RA, Zhao Y, Jones SJ, Marra MA. (2023) A high-throughput pipeline for DNA/RNA/small RNA purification from tissue samples for sequencing. Biotechniques [Epub ahead of print]. [article]