The Neuroscience Monoclonal Antibody Sequencing Initiative (NeuroMabSeq) is a concerted effort to determine and make publicly available hybridoma-derived sequences of monoclonal antibodies (mAbs) valuable to neuroscience research. Over 30 years of research and development efforts including those at the UC Davis/NIH NeuroMab Facility have resulted in the generation of a large collection of mouse mAbs validated for neuroscience research. To enhance dissemination and increase the utility of this valuable resource, researchers at the University of California Davis School of Medicine applied a high-throughput DNA sequencing approach to determine immunoglobulin heavy and light chain variable domain sequences from source hybridoma cells. The resultant set of sequences was made publicly available as a searchable DNA sequence database (neuromabseq.ucdavis.edu) for sharing, analysis and use in downstream applications. The researchers enhanced the utility, transparency, and reproducibility of the existing mAb collection by using these sequences to develop recombinant mAbs. This enabled their subsequent engineering into alternate forms with distinct utility, including alternate modes of detection in multiplexed labeling, and as miniaturized single chain variable fragments or scFvs. The NeuroMabSeq website and database and the corresponding recombinant antibody collection together serve as a public DNA sequence repository of mouse mAb heavy and light chain variable domain sequences and as an open resource for enhancing dissemination and utility of this valuable collection of validated mAbs.
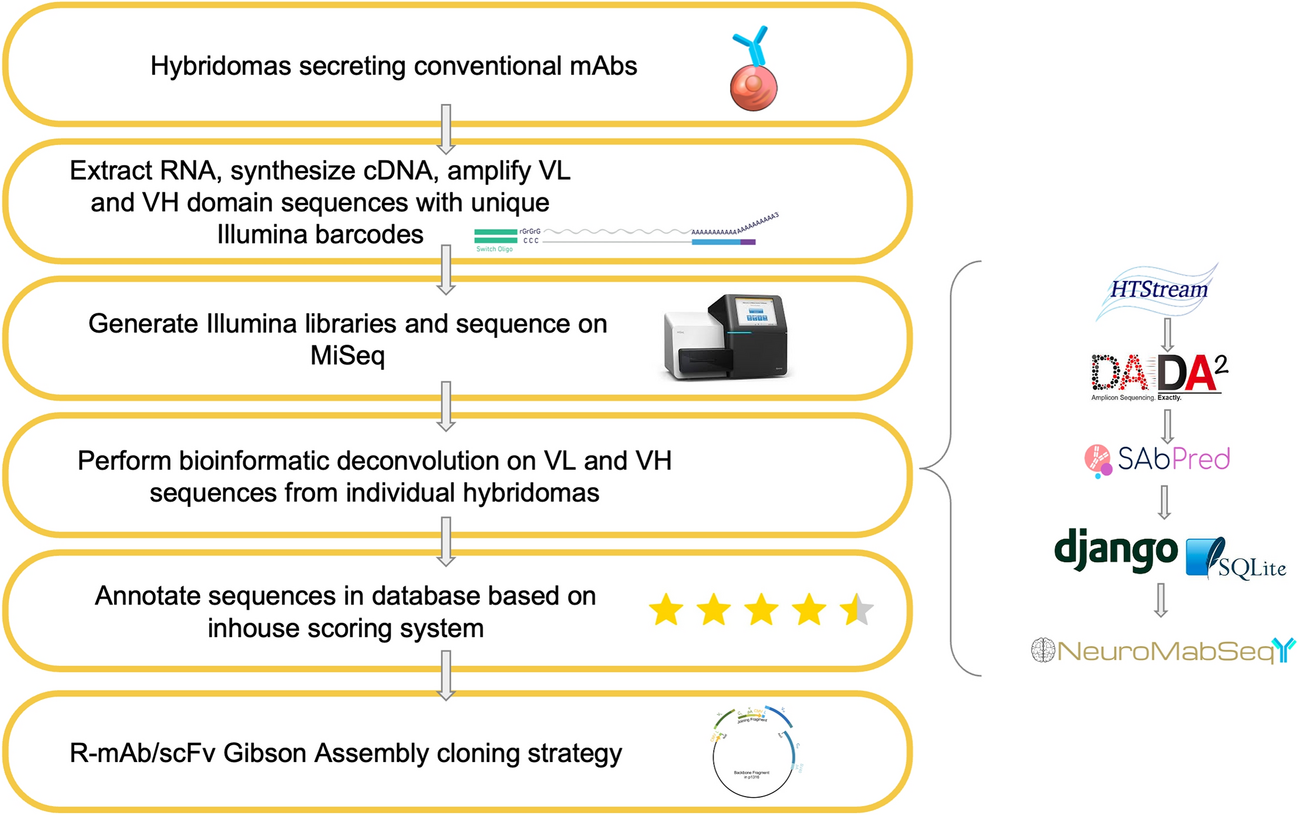
Sequencing workflow and bioinformatics processing
Hybridomas of interest are sequenced using a workflow consisting of RNA extraction, cDNA synthesis, and semi-nested PCR amplification with IgG-specific primers followed by the addition of unique Illumina barcodes to each sample. Illumina libraries are then generated, and adapters are ligated for sequencing on the MiSeq platform. Bioinformatics processing is shown on the right panel. Reads from the Illumina sequencing are run through HTStream for base quality trimming and other read processing. Next, they are passed through DADA2 for amplicon denoising followed by SAbPred ANARCI tool based on the IMGT numbering scheme. All ASVs, metadata, and other quality metrics are uploaded to the NeuroMabSeq database and website where further information and tools are provided to the end users. This includes but is not limited to BlastIR results, BLAT searches across the database, and recommended high quality sequences for recombinant antibody design. Annotations of internally generated scores are provided in addition to other database information. Finally, high quality sequences are used in the design of gene fragments for generation of R-mAb and scFv expression plasmids.
Availability – https://neuromabseq.ucdavis.edu/
Mitchell KG, Gong B, Hunter SS, Burkart-Waco D, Gavira-O’Neill CE, Templeton KM, Goethel ME, Bzymek M, MacNiven LM, Murray KD, Settles ML, Froenicke L, Trimmer JS. (2023) High-volume hybridoma sequencing on the NeuroMabSeq platform enables efficient generation of recombinant monoclonal antibodies and scFvs for neuroscience research. Sci Rep 13(1):16200. [article]