The transcriptional intermediates of RNAs fold into secondary structures with multiple regulatory roles, yet the details of such cotranscriptional RNA folding are largely unresolved in eukaryotes. Researchers at Sun Yat-sen University have developed eSPET-seq (Structural Probing of Elongating Transcripts in eukaryotes), a method to assess the cotranscriptional RNA folding in Saccharomyces cerevisiae. This study reveals pervasive structural transitions during cotranscriptional folding and overall structural similarities between nascent and mature RNAs. Furthermore, a combined analysis with genome-wide R-loop and mutation rate approximations provides quantitative evidence for the antimutator effect of nascent RNA folding through competitive inhibition of the R-loops, known to facilitate transcription-associated mutagenesis. Taken together, these researchers present an experimental evaluation of cotranscriptional folding in eukaryotes and demonstrate the antimutator effect of nascent RNA folding. These results suggest genome-wide coupling between the processing and transmission of genetic information through RNA folding.
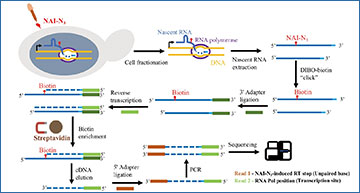
Overview of eSPET-seq procedure and accuracy
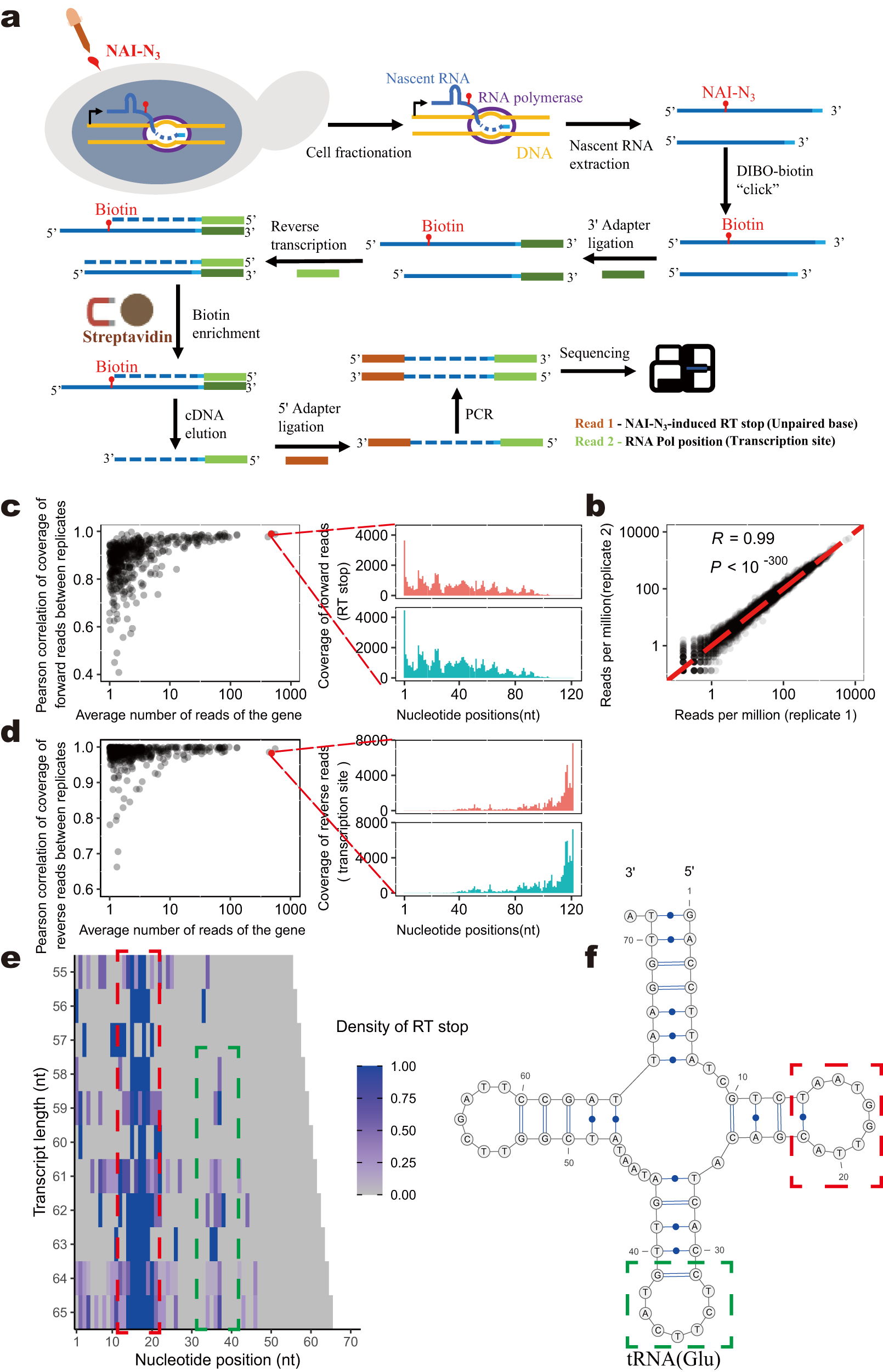
a Experimental procedure of eSPET-seq. b The number of reads mapped to each gene was compared between two biological replicates. Pearson’s correlation is indicated. The dashed red line represents x = y. c The within-gene Pearson’s correlations (y-axis) of the coverage (only the 5′ nucleotide was counted) of the forward reads (representing the RT stops) were calculated between two biological replicates for each gene. The correlation appeared stronger for genes with a higher average number of mapped reads (x-axis). The detailed coverage profile is shown for one sample gene (red dot) on the right. d Similar to (c), except that the correlation calculated for the coverage (only the 5′ nucleotide was counted) of the reverse reads (representing the transcription site) is shown. e Cotranscriptional folding revealed by eSPET-seq. Each row is one transcriptional intermediate of a certain length (y-axis). Each column is a specific nucleotide within the gene. The color of each tile represents the normalized (by 90% winsorization) density of the RT stop, which is indicated by the color scale bar. f The known structure of the yeast glutamate tRNA. The structures outlined by the red and green boxes correspond to the RT density signals outlined by the red and green boxes, respectively, in (e). Source data are provided as a Source Data file.
Availability – Custom R/Python/Perl codes, that were used in data analysis, are available on GitHub (https://github.com/GongwangYu/NascentRNAfolding) and Zenodo (https://zenodo.org/record/8271608)
Yu G, Liu Y, Li Z, Deng S, Wu Z, Zhang X, Chen W, Yang J, Chen X, Yang JR. (2023) Genome-wide probing of eukaryotic nascent RNA structure elucidates cotranscriptional folding and its antimutagenic effect. Nat Commun 14(1):5853. [article]